Application of the Fluorescence-Activating and Absorption-Shifting Tag (FAST) for Flow Cytometry in Methanogenic Archaea
- PMID: 36920214
- PMCID: PMC10132111
- DOI: 10.1128/aem.01786-22
Application of the Fluorescence-Activating and Absorption-Shifting Tag (FAST) for Flow Cytometry in Methanogenic Archaea
Abstract
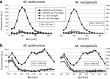
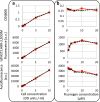
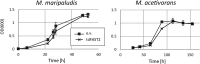
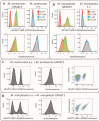
Methane-producing archaea play a crucial role in the global carbon cycle and are used for biotechnological fuel production. Methanogenic model organisms such as Methanococcus maripaludis and Methanosarcina acetivorans have been biochemically characterized and can be genetically engineered by using a variety of existing molecular tools. The anaerobic lifestyle and autofluorescence of methanogens, however, restrict the use of common fluorescent reporter proteins (e.g., GFP and derivatives), which require oxygen for chromophore maturation. Recently, the use of a novel oxygen-independent fluorescent activation and absorption-shifting tag (FAST) was demonstrated with M. maripaludis. Similarly, we now describe the use of the tandem activation and absorption-shifting tag protein 2 (tdFAST2), which fluoresces when the cell-permeable fluorescent ligand (fluorogen) 4-hydroxy-3,5-dimethoxybenzylidene rhodanine (HBR-3,5DOM) is present. Expression of tdFAST2 in M. acetivorans and M. maripaludis is noncytotoxic and tdFAST2:HBR-3,5DOM fluorescence is clearly distinguishable from the autofluorescence. In flow cytometry experiments, mixed methanogen cultures can be distinguished, thereby allowing for the possibility of high-throughput investigations of the characteristic dynamics within single and mixed cultures. IMPORTANCE Methane-producing archaea play an essential role in the global carbon cycle and demonstrate great potential for various biotechnological applications, e.g., biofuel production, carbon dioxide capture, and electrochemical systems. Oxygen sensitivity and high autofluorescence hinder the use of common fluorescent proteins for studying methanogens. By using tdFAST2:HBR-3,5DOM fluorescence, which functions under anaerobic conditions and is distinguishable from the autofluorescence, real-time reporter studies and high-throughput investigation of the mixed culture dynamics of methanogens via flow cytometry were made possible. This will further help accelerate the sustainable exploitation of methanogens.
Keywords: FAST; Methanogenic archaea; Methanosarcina; flow cytometry; fluorescence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bhatia SK, Bhatia RK, Jeon JM, Kumar G, Yang YH. 2019. Carbon dioxide capture and bioenergy production using biological system – A review. Renew Sustain Energy Rev 110:143–158. 10.1016/j.rser.2019.04.070. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

