A Novel CovS Variant Harbored by a Colonization Strain Reduces Streptococcus pyogenes Virulence
- PMID: 36920220
- PMCID: PMC10127592
- DOI: 10.1128/jb.00039-23
A Novel CovS Variant Harbored by a Colonization Strain Reduces Streptococcus pyogenes Virulence
Abstract
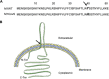
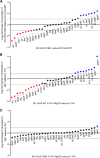
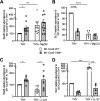
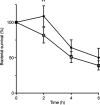
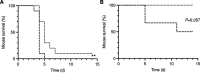
Streptococcus pyogenes, also known as group A Streptococcus, causes a wide variety of diseases ranging from mild noninvasive to severe invasive infections. To identify possible causes of colonization-to-invasive switches, we determined the genomic sequences of 10 isolates from five pairs each composed of an invasive strain and a carriage strain originating from five infectious clusters. Among them, one pair displayed a single-nucleotide difference in covS, encoding the sensor histidine kinase of the two-component CovRS system that controls the expression of 15% of the genome. In contrast to previously described cases where the invasive strains harbor nonfunctional CovS proteins, the carriage strain possessed the mutation covST115C, leading to the replacement of the tyrosine at position 39 by a histidine. The CovSY39H mutation affected the expression of the genes from the CovR regulon in a unique fashion. Genes usually overexpressed in covS mutant strains were underexpressed and vice versa. Furthermore, the covS mutant strain barely responded to the addition of the CovS-signaling compounds Mg2+ and LL-37. The variations in the accumulation of two virulence factors paralleled the transcription modifications. In addition, the covST115C mutant strain showed less survival than its wild-type counterpart in murine macrophages. Finally, in two murine models of infection, the covS mutant strain was less virulent than the wild-type strain. Our study suggests that the CovSY39H protein compromises CovS phosphatase activity and that this yields a noninvasive strain. IMPORTANCE Streptococcus pyogenes, also known as group A Streptococcus, causes a wide variety of diseases, leading to 517,000 deaths yearly. The two-component CovRS system, which responds to MgCl2 and the antimicrobial peptide LL-37, controls the expression of 15% of the genome. Invasive strains may harbor nonfunctional CovS sensor proteins that lead to the derepression of most virulence genes. We isolated a colonization strain that harbors a novel covS mutation. This mutant strain harbored a transcriptome profile opposite that of other covS mutant strains, barely responded to environmental signals, and was less virulent than the wild-type strain. This supports the importance of the derepression of the expression of most virulence genes, via mutations that impact the phosphorylation of the regulator CovR, for favoring S. pyogenes invasive infections.
Keywords: adaptation; covRS; environmental signal; group A Streptococcus; infection; two-component regulatory system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Characterization of a novel covS SNP identified in Australian group A Streptococcus isolates derived from the M1UK lineage.mBio. 2025 Feb 5;16(2):e0336624. doi: 10.1128/mbio.03366-24. Epub 2024 Dec 17. mBio. 2025. PMID: 39688411 Free PMC article.
-
A natural inactivating mutation in the CovS component of the CovRS regulatory operon in a pattern D Streptococcal pyogenes strain influences virulence-associated genes.J Biol Chem. 2013 Mar 1;288(9):6561-73. doi: 10.1074/jbc.M112.442657. Epub 2013 Jan 13. J Biol Chem. 2013. PMID: 23316057 Free PMC article.
-
CovRS-Regulated Transcriptome Analysis of a Hypervirulent M23 Strain of Group A Streptococcus pyogenes Provides New Insights into Virulence Determinants.J Bacteriol. 2015 Oct;197(19):3191-205. doi: 10.1128/JB.00511-15. Epub 2015 Jul 27. J Bacteriol. 2015. PMID: 26216843 Free PMC article.
-
The two faces of Janus: virulence gene regulation by CovR/S in group A streptococci.Mol Microbiol. 2007 Apr;64(1):34-41. doi: 10.1111/j.1365-2958.2007.05649.x. Mol Microbiol. 2007. PMID: 17376070 Review.
-
Cellular interactions of covR/S mutant group A Streptococci.Microbes Infect. 2018 Oct-Nov;20(9-10):531-535. doi: 10.1016/j.micinf.2017.12.009. Epub 2017 Dec 26. Microbes Infect. 2018. PMID: 29287985 Review.
Cited by
-
The double-edged role of FASII regulator FabT in Streptococcus pyogenes infection.Nat Commun. 2024 Oct 4;15(1):8593. doi: 10.1038/s41467-024-52637-3. Nat Commun. 2024. PMID: 39366941 Free PMC article.
-
A Streptococcus pyogenes DegV protein regulates the membrane lipid content and limits the formation of extracellular vesicles.PLoS One. 2023 Apr 27;18(4):e0284402. doi: 10.1371/journal.pone.0284402. eCollection 2023. PLoS One. 2023. PMID: 37104252 Free PMC article.
-
Clinical Snapshot of Group A Streptococcal Isolates from an Australian Tertiary Hospital.Pathogens. 2024 Nov 1;13(11):956. doi: 10.3390/pathogens13110956. Pathogens. 2024. PMID: 39599509 Free PMC article.
-
Characterization of a novel covS SNP identified in Australian group A Streptococcus isolates derived from the M1UK lineage.mBio. 2025 Feb 5;16(2):e0336624. doi: 10.1128/mbio.03366-24. Epub 2024 Dec 17. mBio. 2025. PMID: 39688411 Free PMC article.
-
Reduced interleukin-18 secretion by human monocytic cells in response to infections with hyper-virulent Streptococcus pyogenes.J Biomed Sci. 2024 Feb 27;31(1):26. doi: 10.1186/s12929-024-01014-9. J Biomed Sci. 2024. PMID: 38408992 Free PMC article.
References
-
- Li Z, Sakota V, Jackson D, Franklin AR, Beall B. Active Bacterial Core Surveillance/Emerging Infections Program Network. 2003. Array of M protein gene subtypes in 1064 recent invasive group A streptococcus isolates recovered from the active bacterial core surveillance. J Infect Dis 188:1587–1592. 10.1086/379050. - DOI - PubMed

