Population genetics and geographic origins of mallards harvested in northwestern Ohio
- PMID: 36920978
- PMCID: PMC10016643
- DOI: 10.1371/journal.pone.0282874
Population genetics and geographic origins of mallards harvested in northwestern Ohio
Abstract
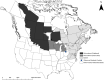
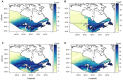
The genetic composition of mallards in eastern North America has been changed by release of domestically-raised, game-farm mallards to supplement wild populations for hunting. We sampled 296 hatch-year mallards harvested in northwestern Ohio, October-December 2019. The aim was to determine their genetic ancestry and geographic origin to understand the geographic extent of game-farm mallard introgression into wild populations in more westward regions of North America. We used molecular analysis to detect that 35% of samples were pure wild mallard, 12% were early generation hybrids between wild and game-farm mallards (i.e., F1-F3), and the remaining 53% of samples were assigned as part of a hybrid swarm. Percentage of individuals in our study with some form of hybridization with game-farm mallard (65%) was greater than previously detected farther south in the mid-continent (~4%), but less than the Atlantic coast of North America (~ 92%). Stable isotope analysis using δ2Hf suggested that pure wild mallards originated from areas farther north and west than hybrid mallards. More specifically, 17% of all Ohio samples had δ2Hf consistent with more western origins in the prairies, parkland, or boreal regions of the mid-continent of North America, with 55%, 35%, and 10% of these being genetically wild, hybrid swarm, and F3, respectively. We conclude that continued game-farm introgression into wild mallards is not isolated to the eastern population of mallards in North America, and may be increasing and more widespread than previously detected. Mallards in our study had greater incidence of game-farm hybridization than other locales in the mid-continent but less than eastern North American regions suggesting further need to understand game-farm mallard genetic variation and movement across the continent.
Copyright: © 2023 Schummer et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Kiple KF. The Cambridge world history of food. Cambridge University Press. 2001. Nov 22.
-
- Mock KE, Latch EK, Rhodes OE. Assessing losses of genetic diversity due to translocation: long-term case histories in Merriam’s turkey (Meleagris gallopavo merriami). Conserv Genet. 2004;5: 631–645.
-
- Reynolds M and Klavitter J. Translocation of wild Laysan duck to establish a population at Midway Atoll National Wildlife Refuge, United States and US Pacific Flyway. Conserv Evid. 2006;3: 6–8.
-
- Rodríguez-Clark KM, Maldonado JE, Ascanio D, Gamero E, Ovalle Moleiro L, Pérez-Emán J, et al.. Using genetics to understand and conserve the Red Siskin (Carduelis cucullata). J of the Nat Finch and Softbill Soc. 2011;1:25–30.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

