A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance
- PMID: 36921044
- PMCID: PMC10017050
- DOI: 10.1126/sciadv.ade2175
A hotspot for posttranslational modifications on the androgen receptor dimer interface drives pathology and anti-androgen resistance
Abstract
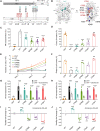
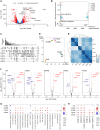
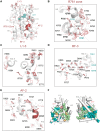
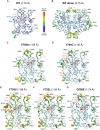
Mutations of the androgen receptor (AR) associated with prostate cancer and androgen insensitivity syndrome may profoundly influence its structure, protein interaction network, and binding to chromatin, resulting in altered transcription signatures and drug responses. Current structural information fails to explain the effect of pathological mutations on AR structure-function relationship. Here, we have thoroughly studied the effects of selected mutations that span the complete dimer interface of AR ligand-binding domain (AR-LBD) using x-ray crystallography in combination with in vitro, in silico, and cell-based assays. We show that these variants alter AR-dependent transcription and responses to anti-androgens by inducing a previously undescribed allosteric switch in the AR-LBD that increases exposure of a major methylation target, Arg761. We also corroborate the relevance of residues Arg761 and Tyr764 for AR dimerization and function. Together, our results reveal allosteric coupling of AR dimerization and posttranslational modifications as a disease mechanism with implications for precision medicine.
Figures




References
-
- W. D. Tilley, C. M. Wilson, M. Marcelli, M. J. McPhaul, Androgen receptor gene expression in human prostate carcinoma cell lines. Cancer Res. 17, 5382–5386 (1990). - PubMed
-
- A. Jiménez-Panizo, P. Pérez, A. M. Rojas, P. Fuentes-Prior, E. Estébanez-Perpiñá, Non-canonical dimerization of the androgen receptor and other nuclear receptors: Implications for human disease. Endocr. Relat. Cancer 26, R479–R497 (2019). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

