Discovering highly potent antimicrobial peptides with deep generative model HydrAMP
- PMID: 36922490
- PMCID: PMC10017685
- DOI: 10.1038/s41467-023-36994-z
Discovering highly potent antimicrobial peptides with deep generative model HydrAMP
Erratum in
-
Author Correction: Discovering highly potent antimicrobial peptides with deep generative model HydrAMP.Nat Commun. 2023 Aug 23;14(1):5129. doi: 10.1038/s41467-023-40879-6. Nat Commun. 2023. PMID: 37612279 Free PMC article. No abstract available.
Abstract
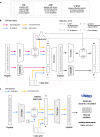
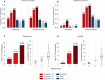
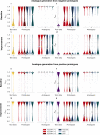
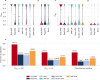
Antimicrobial peptides emerge as compounds that can alleviate the global health hazard of antimicrobial resistance, prompting a need for novel computational approaches to peptide generation. Here, we propose HydrAMP, a conditional variational autoencoder that learns lower-dimensional, continuous representation of peptides and captures their antimicrobial properties. The model disentangles the learnt representation of a peptide from its antimicrobial conditions and leverages parameter-controlled creativity. HydrAMP is the first model that is directly optimized for diverse tasks, including unconstrained and analogue generation and outperforms other approaches in these tasks. An additional preselection procedure based on ranking of generated peptides and molecular dynamics simulations increases experimental validation rate. Wet-lab experiments on five bacterial strains confirm high activity of nine peptides generated as analogues of clinically relevant prototypes, as well as six analogues of an inactive peptide. HydrAMP enables generation of diverse and potent peptides, making a step towards resolving the antimicrobial resistance crisis.
© 2023. The Author(s).
Conflict of interest statement
The authors P.Sz., M.Mo., T.G., R.J., M.B., D.N., M.Mi, P.Se, W.K, and E.S have issued a patent for the newly generated AMPs under the patent number P.443243. The other authors declare no competing interests.
Figures




References
-
- CDC. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA, USA: US Department of Health and Human Services, CDC (2019).
-
- O’Neill, J. Tackling drug-resistant infections globally: final report and recommendations. The Review on Antimicrobial Resistance, Government of the United Kingdom (2016).
-
- Magana M. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020;20:e216–e230. - PubMed
-
- Czaplewski L, et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016;16:239–251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

