Loss of MBNL1-mediated retrograde BDNF signaling in the myotonic dystrophy brain
- PMID: 36922901
- PMCID: PMC10018927
- DOI: 10.1186/s40478-023-01540-x
Loss of MBNL1-mediated retrograde BDNF signaling in the myotonic dystrophy brain
Abstract
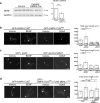
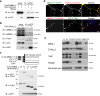
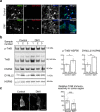
Reduced brain volume including atrophy in grey and white matter is commonly seen in myotonic dystrophy type 1 (DM1). DM1 is caused by an expansion of CTG trinucleotide repeats in the 3' untranslated region (UTR) of the Dystrophia Myotonica Protein Kinase (DMPK) gene. Mutant DMPK mRNA containing expanded CUG RNA (DMPK-CUGexp) sequesters cytoplasmic MBNL1, resulting in morphological impairment. How DMPK-CUGexp and loss of MBNL1 cause histopathological phenotypes in the DM1 brain remains elusive. Here, we show that BDNF-TrkB retrograde transport is impaired in neurons expressing DMPK-CUGexp due to loss of cytoplasmic MBNL1 function. We reveal that mature BDNF protein levels are reduced in the brain of the DM1 mouse model EpA960/CaMKII-Cre. Exogenous BDNF treatment did not rescue impaired neurite outgrowth in neurons expressing DMPK-CUGexp, whereas overexpression of the cytoplasmic MBNL1 isoform in DMPK-CUGexp-expressing neurons improved their responsiveness to exogenous BDNF. We identify dynein light chain LC8-type 2, DYNLL2, as an MBNL1-interacting protein and demonstrate that their interaction is RNA-independent. Using time-lapse imaging, we show that overexpressed MBNL1 and DYNLL2 move along axonal processes together and that MBNL1-knockdown impairs the motility of mCherry-tagged DYNLL2, resulting in a reduced percentage of retrograde DYNLL2 movement. Examination of the distribution of DYNLL2 and activated phospho-TrkB (pTrkB) receptor in EpA960/CaMKII-Cre brains revealed an increase in the postsynaptic membrane fraction (LP1), indicating impaired retrograde transport. Finally, our neuropathological analysis of postmortem DM1 tissue reveals that reduced cytoplasmic MBNL1 expression is associated with an increase in DYNLL2 and activated pTrkB receptor levels in the synaptosomal fraction. Together, our results support that impaired MBNL1-mediated retrograde BDNF-TrkB signaling may contribute to the histopathological phenotypes of DM1.
Keywords: BDNF; DYNLL2; MBNL1; Myotonic dystrophy; Retrograde transport.
© 2023. The Author(s).
Conflict of interest statement
The authors have no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

