Whole-exome Sequencing of Nigerian Prostate Tumors from the Prostate Cancer Transatlantic Consortium (CaPTC) Reveals DNA Repair Genes Associated with African Ancestry
- PMID: 36922933
- PMCID: PMC10010347
- DOI: 10.1158/2767-9764.CRC-22-0136
Whole-exome Sequencing of Nigerian Prostate Tumors from the Prostate Cancer Transatlantic Consortium (CaPTC) Reveals DNA Repair Genes Associated with African Ancestry
Abstract
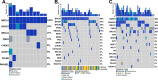
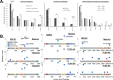
In this study, we used whole-exome sequencing of a cohort of 45 advanced-stage, treatment-naïve Nigerian (NG) primary prostate cancer tumors and 11 unmatched nontumor tissues to compare genomic mutations with African American (AA) and European American (EA) The Cancer Genome Atlas (TCGA) prostate cancer. NG samples were collected from six sites in central and southwest Nigeria. After whole-exome sequencing, samples were processed using GATK best practices. BRCA1 (100%), BARD1 (45%), BRCA2 (27%), and PMS2(18%) had germline alterations in at least two NG nontumor samples. Across 111 germline variants, the AA cohort reflected a pattern [BRCA1 (68%), BARD1 (34%), BRCA2 (28%), and PMS2 (16%)] similar to NG samples. Of the most frequently mutated genes, BRCA1 showed a statistically (P ≤ 0.05) higher germline mutation frequency in men of African ancestry (MAA) and increasing variant frequency with increased African ancestry. Disaggregating gene-level mutation frequencies by variants revealed both ancestry-linked and NG-specific germline variant patterns. Driven by rs799917 (T>C), BRCA1 showed an increasing mutation frequency as African ancestry increased. BRCA2_rs11571831 was present only in MAA, and BRCA2_rs766173 was elevated in NG men. A total of 133 somatic variants were present in 26 prostate cancer-associated genes within the NG tumor cohort. BRCA2 (27%), APC (20%), ATM (20%), BRCA1 (13%), DNAJC6 (13%), EGFR (13%), MAD1L1 (13%), MLH1 (11%), and PMS2 (11%) showed mutation frequencies >10%. Compared with TCGA cohorts, NG tumors showed statistically significant elevated frequencies of BRCA2, APC, and BRCA1. The NG cohort variant pattern shared similarities (cosign similarities ≥0.734) with Catalogue of Somatic Mutations in Cancer signatures 5 and 6, and mutated genes showed significant (q < 0.001) gene ontology (GO) and functional enrichment in mismatch repair and non-homologous repair deficiency pathways. Here, we showed that mutations in DNA damage response genes were higher in NG prostate cancer samples and that a portion of those mutations correlate with African ancestry. Moreover, we identified variants of unknown significance that may contribute to population-specific routes of tumorigenesis and treatment. These results present the most comprehensive characterization of the NG prostate cancer exome to date and highlight the need to increase diversity of study populations.
Significance: MAA have higher rates of prostate cancer incidence and mortality, however, are severely underrepresented in genomic studies. This is the first study utilizing whole-exome sequencing in NG men to identify West African ancestry-linked variant patterns that impact DNA damage repair pathways.
© 2022 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
F. Mohammed reports other from BIO Ventures for Global Health outside the submitted work. M. Davis reports grants from Weill Cornell Prostate SPORE during the conduct of the study; grants from Genentech and grants and non-financial support from NIH outside the submitted work. P. Polak reports other from C2i Genomcis outside the submitted work. C. Yates reports other from Leidos contract YT16-010 (Project ID# 001.050.0010), NCI contracts HHSN261201600732P (University of Florida); grants from U54-MD007585-26, U54 CA118623 (NIH/NCI), and PC170315P1, W81XWH-18-1-0589) awarded to C. Yates during the conduct of the study; personal fees from Riptide Biosciences, QED Therapeutics, and Amgen and other from Riptide Biosciences outside the submitted work. No disclosures were reported by the other authors.
Figures


References
-
- Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144:1941–53. - PubMed
-
- Jensen OM, Storm HH. Cancer registration: principles and methods. Reporting of results. IARC Sci Publ 1991;108–25. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

