A Spatially Resolved Mechanistic Growth Law for Cancer Drug Development Predicting Tumor Growing Fractions
- PMID: 36923310
- PMCID: PMC10010375
- DOI: 10.1158/2767-9764.CRC-22-0032
A Spatially Resolved Mechanistic Growth Law for Cancer Drug Development Predicting Tumor Growing Fractions
Abstract
Mathematical models used in preclinical drug discovery tend to be empirical growth laws. Such models are well suited to fitting the data available, mostly longitudinal studies of tumor volume; however, they typically have little connection with the underlying physiologic processes. This lack of a mechanistic underpinning restricts their flexibility and potentially inhibits their translation across studies including from animal to human. Here we present a mathematical model describing tumor growth for the evaluation of single-agent cytotoxic compounds that is based on mechanistic principles. The model can predict spatial distributions of cell subpopulations and account for spatial drug distribution effects within tumors. Importantly, we demonstrate that the model can be reduced to a growth law similar in form to the ones currently implemented in pharmaceutical drug development for preclinical trials so that it can integrated into the current workflow. We validate this approach for both cell-derived xenograft and patient-derived xenograft (PDX) data. This shows that our theoretical model fits as well as the best performing and most widely used models. However, in addition, the model is also able to accurately predict the observed growing fraction of tumours. Our work opens up current preclinical modeling studies to also incorporating spatially resolved and multimodal data without significant added complexity and creates the opportunity to improve translation and tumor response predictions.
Significance: This theoretical model has the same mathematical structure as that currently used for drug development. However, its mechanistic basis enables prediction of growing fraction and spatial variations in drug distribution.
© 2022 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
A. Nasim reports grants from AstraZeneca during the conduct of the study. No disclosures were reported by the other authors.
Figures
 and
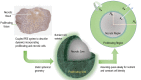
and  being the necrotic and total tumor radii, respectively. (Histologic image—a day-24 CDX xenograft with Ki-67 stain.)
being the necrotic and total tumor radii, respectively. (Histologic image—a day-24 CDX xenograft with Ki-67 stain.)

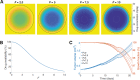
 . C, Tumor volume (blue) and growth fraction (red) dynamics for dosing strength fixed at 50 mg/kg dosed on days 1, 8, 15, for
. C, Tumor volume (blue) and growth fraction (red) dynamics for dosing strength fixed at 50 mg/kg dosed on days 1, 8, 15, for  (blue: bottom to top curves, respectively, red: top to bottom curves, respectively).
(blue: bottom to top curves, respectively, red: top to bottom curves, respectively).  describes full drug penetration, with increasing
describes full drug penetration, with increasing  corresponding to a reduction in drug penetration. At
corresponding to a reduction in drug penetration. At  , the drug effect is largely restricted to the surface of the tumor.
, the drug effect is largely restricted to the surface of the tumor.References
-
- Yates JWT, Byrne H, Chapman SC, Chen T, Cucurull-Sanchez L, Delgado-Sanmartin J, et al. . Opportunities for quantitative translational modeling in oncology. Clin Pharmacol Ther 2020;108:447–57. - PubMed
-
- Simeoni M, Magni P, Cammia C, De Nicolao G, Croci V, Pesenti E, et al. . Predictive pharmacokinetic-pharmacodynamic modeling of tumor growth kinetics in xenograft models after administration of anticancer agents. Cancer Res 2004;64:1094–101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

