Primary refractory plasmablastic lymphoma: A precision oncology approach
- PMID: 36923431
- PMCID: PMC10008852
- DOI: 10.3389/fonc.2023.1129405
Primary refractory plasmablastic lymphoma: A precision oncology approach
Abstract
Introduction: Hematologic malignancies are currently underrepresented in multidisciplinary molecular-tumor-boards (MTB). This study assesses the potential of precision-oncology in primary-refractory plasmablastic-lymphoma (prPBL), a highly lethal blood cancer.
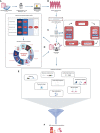
Methods: We evaluated clinicopathological and molecular-genetic data of 14 clinically annotated prPBL-patients from initial diagnosis. For this proof-of-concept study, we employed our certified institutional MTB-pipeline (University-Cancer-Center-Schleswig-Holstein, UCCSH) to annotate a comprehensive dataset within the scope of a virtual MTB-setting, ultimately recommending molecularly stratified therapies. Evidence-levels for MTB-recommendations were defined in accordance with the NCT/DKTK and ESCAT criteria.
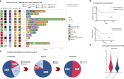
Results: Median age in the cohort was 76.5 years (range 56-91), 78.6% of patients were male, 50% were HIV-positive and clinical outcome was dismal. Comprehensive genomic/transcriptomic analysis revealed potential recommendations of a molecularly stratified treatment option with evidence-levels according to NCT/DKTK of at least m2B/ESCAT of at least IIIA were detected for all 14 prPBL-cases. In addition, immunohistochemical-assessment (CD19/CD30/CD38/CD79B) revealed targeted treatment-recommendations in all 14 cases. Genetic alterations were classified by treatment-baskets proposed by Horak et al. Hereby, we identified tyrosine-kinases (TK; n=4), PI3K-MTOR-AKT-pathway (PAM; n=3), cell-cycle-alterations (CC; n=2), RAF-MEK-ERK-cascade (RME; n=2), immune-evasion (IE; n=2), B-cell-targets (BCT; n=25) and others (OTH; n=4) for targeted treatment-recommendations. The minimum requirement for consideration of a drug within the scope of the study was FDA-fast-track development.
Discussion: The presented proof-of-concept study demonstrates the clinical potential of precision-oncology, even in prPBL-patients. Due to the aggressive course of the disease, there is an urgent medical-need for personalized treatment approaches, and this population should be considered for MTB inclusion at the earliest time.
Keywords: molecular tumor board; plasmablastic lymphoma; recurrent aberrations; targeted therapy; whole exome sequencing; whole transcriptome sequencing.
Copyright © 2023 Witte, Fähnrich, Künstner, Riedl, Fliedner, Reimer, Hertel, von Bubnoff, Bernard, Merz, Busch, Feller and Gebauer.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Subbiah V, Puzanov I, Blay JY, Chau I, Lockhart AC, Raje NS, et al. Pan-cancer efficacy of vemurafenib in BRAF (V600)-mutant non-melanoma cancers. Cancer Discovery (2020) 10(5):657–63. doi: 10.1158/2159-8290.CD-19-1265 - DOI - PMC - PubMed
-
- Horak P, Heining C, Kreutzfeldt S, Hutter B, Mock A, Hullein J, et al. Comprehensive genomic and transcriptomic analysis for guiding therapeutic decisions in patients with rare cancers. Cancer Discovery (2021) 11(11):2780–95. doi: 10.1158/2159-8290.CD-21-0126 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

