Identification and validation of metabolism-related hub genes in idiopathic pulmonary fibrosis
- PMID: 36923791
- PMCID: PMC10010493
- DOI: 10.3389/fgene.2023.1058582
Identification and validation of metabolism-related hub genes in idiopathic pulmonary fibrosis
Abstract
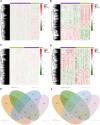
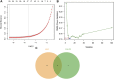
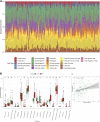
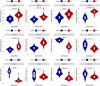
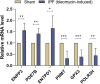
Background: Idiopathic pulmonary fibrosis (IPF) is a fatal and irreversible interstitial lung disease. The specific mechanisms involved in the pathogenesis of IPF are not fully understood, while metabolic dysregulation has recently been demonstrated to contribute to IPF. This study aims to identify key metabolism-related genes involved in the progression of IPF, providing new insights into the pathogenesis of IPF. Methods: We downloaded four datasets (GSE32537, GSE110147, GSE150910, and GSE92592) from the Gene Expression Omnibus (GEO) database and identified differentially expressed metabolism-related genes (DEMRGs) in lung tissues of IPF by comprehensive analysis. Then, we performed GO, KEGG, and Reactome enrichment analyses of the DEMRGs. Subsequently, key DEMRGs were identified by machine-learning algorithms. Next, miRNAs regulating these key DEMRGs were predicted by integrating the GSE32538 (IPF miRNA dataset) and the miRWalk database. The Cytoscape software was used to visualize miRNA-mRNA regulatory networks. In addition, the relative levels of immune cells were assessed by the CIBERSORT algorithm, and the correlation of key DEMRGs with immune cells was calculated. Finally, the mRNA expression of the key DEMRGs was validated in two external independent datasets and an in vivo experiment. Results: A total of 101 DEMRGs (51 upregulated and 50 downregulated) were identified. Six key DEMRGs (ENPP3, ENTPD1, GPX3, PDE7B, PNMT, and POLR3H) were further identified using two machine-learning algorithms (LASSO and SVM-RFE). In the lung tissue of IPF patients, the expression levels of ENPP3, ENTPD1, and PDE7B were upregulated, and the expression levels of GPX3, PNMT, and POLR3H were downregulated. In addition, the miRNA-mRNA regulatory network of key DEMRGs was constructed. Then, the expression levels of key DEMRGs were validated in two independent external datasets (GSE53845 and GSE213001). Finally, we verified the key DEMRGs in the lung tissue of bleomycin-induced pulmonary fibrosis mice by qRT-PCR. Conclusion: Our study identified key metabolism-related genes that are differentially expressed in the lung tissue of IPF patients. Our study emphasizes the critical role of metabolic dysregulation in IPF, offers potential therapeutic targets, and provides new insights for future studies.
Keywords: IPF; bioinformatics; biomarker; differentially expressed genes; gene expression omnibus; hub genes; metabolic; metabolism.
Copyright © 2023 Zeng, Huang, Guo, Cao, Yang and Ouyang.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

