The Emerging Role of Super-enhancers as Therapeutic Targets in The Digestive System Tumors
- PMID: 36923930
- PMCID: PMC10008685
- DOI: 10.7150/ijbs.78535
The Emerging Role of Super-enhancers as Therapeutic Targets in The Digestive System Tumors
Abstract
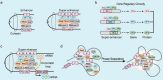
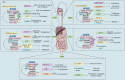
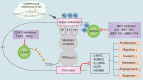
Digestive system tumors include malignancies of the stomach, pancreas, colon, rectum, and the esophagus, and are associated with high morbidity and mortality. Aberrant epigenetic modifications play a vital role in the progression of digestive system tumors. The aberrant transcription of key oncogenes is driven by super-enhancers (SEs), which are characterized by large clusters of enhancers with significantly high density of transcription factors, cofactors, and epigenetic modulatory proteins. The SEs consist of critical epigenetic regulatory elements, which modulate the biological characteristics of digestive system tumors including tumor cell identity and differentiation, tumorigenesis, environmental response, immune response, and chemotherapeutic resistance. The core transcription regulatory loop of the digestive system tumors is complex and a high density of transcription regulatory complexes in the SEs and the crosstalk between SEs and the noncoding RNAs. In this review, we summarized the known characteristics and functions of the SEs in the digestive system tumors. Furthermore, we discuss the oncogenic roles and regulatory mechanisms of SEs in the digestive system tumors. We highlight the role of SE-driven genes, enhancer RNAs (eRNAs), lncRNAs, and miRNAs in the digestive system tumor growth and progression. Finally, we discuss clinical significance of the CRISPR-Cas9 gene editing system and inhibitors of SE-related proteins such as BET and CDK7 as potential cancer therapeutics.
Keywords: digestive system tumors; non-coding RNAs; super-enhancer; therapeutic targets.; tumor progression.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
References
-
- Nagaraju GP, Kasa P, Dariya B, Surepalli N, Peela S, Ahmad S. Epigenetics and therapeutic targets in gastrointestinal malignancies. Drug Discov Today. 2021. - PubMed
-
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

