Extreme differences in SARS-CoV-2 viral loads among respiratory specimen types during presumed pre-infectious and infectious periods
- PMID: 36926220
- PMCID: PMC10013338
- DOI: 10.1093/pnasnexus/pgad033
Extreme differences in SARS-CoV-2 viral loads among respiratory specimen types during presumed pre-infectious and infectious periods
Abstract
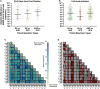
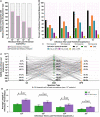
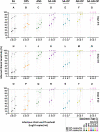
SARS-CoV-2 viral-load measurements from a single-specimen type are used to establish diagnostic strategies, interpret clinical-trial results for vaccines and therapeutics, model viral transmission, and understand virus-host interactions. However, measurements from a single-specimen type are implicitly assumed to be representative of other specimen types. We quantified viral-load timecourses from individuals who began daily self-sampling of saliva, anterior-nares (nasal), and oropharyngeal (throat) swabs before or at the incidence of infection with the Omicron variant. Viral loads in different specimen types from the same person at the same timepoint exhibited extreme differences, up to 109 copies/mL. These differences were not due to variation in sample self-collection, which was consistent. For most individuals, longitudinal viral-load timecourses in different specimen types did not correlate. Throat-swab and saliva viral loads began to rise as many as 7 days earlier than nasal-swab viral loads in most individuals, leading to very low clinical sensitivity of nasal swabs during the first days of infection. Individuals frequently exhibited presumably infectious viral loads in one specimen type while viral loads were low or undetectable in other specimen types. Therefore, defining an individual as infectious based on assessment of a single-specimen type underestimates the infectious period, and overestimates the ability of that specimen type to detect infectious individuals. For diagnostic COVID-19 testing, these three single-specimen types have low clinical sensitivity, whereas a combined throat-nasal swab, and assays with high analytical sensitivity, was inferred to have significantly better clinical sensitivity to detect presumed pre-infectious and infectious individuals.
Keywords: COVID-19; testing strategies; viral loads.
© The Author(s) 2023. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures



References
-
- NIH . 2021. Therapeutic Management of Adults With COVID-19. https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/.
LinkOut - more resources
Full Text Sources
Miscellaneous

