Before and after AlphaFold2: An overview of protein structure prediction
- PMID: 36926275
- PMCID: PMC10011655
- DOI: 10.3389/fbinf.2023.1120370
Before and after AlphaFold2: An overview of protein structure prediction
Abstract
Three-dimensional protein structure is directly correlated with its function and its determination is critical to understanding biological processes and addressing human health and life science problems in general. Although new protein structures are experimentally obtained over time, there is still a large difference between the number of protein sequences placed in Uniprot and those with resolved tertiary structure. In this context, studies have emerged to predict protein structures by methods based on a template or free modeling. In the last years, different methods have been combined to overcome their individual limitations, until the emergence of AlphaFold2, which demonstrated that predicting protein structure with high accuracy at unprecedented scale is possible. Despite its current impact in the field, AlphaFold2 has limitations. Recently, new methods based on protein language models have promised to revolutionize the protein structural biology allowing the discovery of protein structure and function only from evolutionary patterns present on protein sequence. Even though these methods do not reach AlphaFold2 accuracy, they already covered some of its limitations, being able to predict with high accuracy more than 200 million proteins from metagenomic databases. In this mini-review, we provide an overview of the breakthroughs in protein structure prediction before and after AlphaFold2 emergence.
Keywords: AlphaFold; free modeling; protein language model; protein structure prediction; template-based modeling.
Copyright © 2023 Bertoline, Lima, Krieger and Teixeira.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
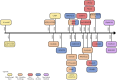
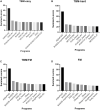
References
-
- Agnihotry S., Pathak R. K., Singh D. B., Tiwari A., Hussain I. (2022). “Protein structure prediction,” in Bioinformatics (Amsterdam, Netherlands: Elsevier; ), 177–188. 10.1016/B978-0-323-89775-4.00023-7 - DOI
Publication types
LinkOut - more resources
Full Text Sources

