A phylogenetic study of dengue virus in urban Vietnam shows long-term persistence of endemic strains
- PMID: 36926448
- PMCID: PMC10013730
- DOI: 10.1093/ve/vead012
A phylogenetic study of dengue virus in urban Vietnam shows long-term persistence of endemic strains
Abstract
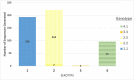
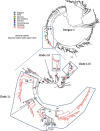
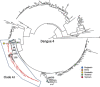
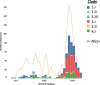
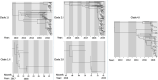
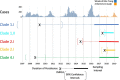
Dengue virus (DENV) causes repeated outbreaks of disease in endemic areas, with patterns of local transmission strongly influenced by seasonality, importation via human movement, immunity, and vector control efforts. An understanding of how each of these interacts to enable endemic transmission (continual circulation of local virus strains) is largely unknown. There are times of the year when no cases are reported, often for extended periods of time, perhaps wrongly implying the successful eradication of a local strain from that area. Individuals who presented at a clinic or hospital in four communes in Nha Trang, Vietnam, were initially tested for DENV antigen presence. Enrolled positive individuals then had their corresponding household members invited to participate, and those who enrolled were tested for DENV. The presence of viral nucleic acid in all samples was confirmed using quantitative polymerase chain reaction, and positive samples were then whole-genome sequenced using an amplicon and target enrichment library preparation techniques and Illumina MiSeq sequencing technology. Generated consensus genome sequences were then analysed using phylogenetic tree reconstruction to categorise sequences into clades with a common ancestor, enabling investigations of both viral clade persistence and introductions. Hypothetical introduction dates were additionally assessed using a molecular clock model that calculated the time to the most recent common ancestor (TMRCA). We obtained 511 DENV whole-genome sequences covering four serotypes and more than ten distinct viral clades. For five of these clades, we had sufficient data to show that the same viral lineage persisted for at least several months. We noted that some clades persisted longer than others during the sampling time, and by comparison with other published sequences from elsewhere in Vietnam and around the world, we saw that at least two different viral lineages were introduced into the population during the study period (April 2017-2019). Next, by inferring the TMRCA from the construction of molecular clock phylogenies, we predicted that two of the viral lineages had been present in the study population for over a decade. We observed five viral lineages co-circulating in Nha Trang from three DENV serotypes, with two likely to have remained as uninterrupted transmission chains for a decade. This suggests clade cryptic persistence in the area, even during periods of low reported incidence.
Keywords: dengue; persistence; phylogenetics; pransmission; sequencing; virus.
© The Author(s) 2023. Published by Oxford University Press.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

