Intestinal bacteria and colorectal cancer: etiology and treatment
- PMID: 36927206
- PMCID: PMC10026918
- DOI: 10.1080/19490976.2023.2185028
Intestinal bacteria and colorectal cancer: etiology and treatment
Abstract
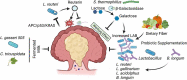
The etiology of colorectal cancer (CRC) is influenced by bacterial communities that colonize the gastrointestinal tract. These microorganisms derive essential nutrients from indigestible dietary or host-derived compounds and activate molecular signaling pathways necessary for normal tissue and immune function. Associative and mechanistic studies have identified bacterial species whose presence may increase CRC risk, including notable examples such as Fusobacterium nucleatum, Enterotoxigenic Bacteroides fragilis, and pks+ E. coli. In recent years this work has expanded in scope to include aspects of host mutational status, intra-tumoral microbial heterogeneity, transient infection, and the cumulative influence of multiple carcinogenic bacteria after sequential or co-colonization. In this review, we will provide an updated overview of how host-bacteria interactions influence CRC development, how this knowledge may be utilized to diagnose or prevent CRC, and how the gut microbiome influences CRC treatment efficacy.
Keywords: Colorectal cancer; genotoxin; immunotherapy; inflammation; microbiome; probiotics.
Conflict of interest statement
The authors report they have no competing interests to declare.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
