Quinolinate promotes macrophage-induced immune tolerance in glioblastoma through the NMDAR/PPARγ signaling axis
- PMID: 36927729
- PMCID: PMC10020159
- DOI: 10.1038/s41467-023-37170-z
Quinolinate promotes macrophage-induced immune tolerance in glioblastoma through the NMDAR/PPARγ signaling axis
Abstract
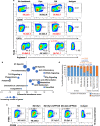
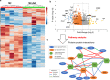
There has been considerable scientific effort dedicated to understanding the biologic consequence and therapeutic implications of aberrant tryptophan metabolism in brain tumors and neurodegenerative diseases. A majority of this work has focused on the upstream metabolism of tryptophan; however, this has resulted in limited clinical application. Using global metabolomic profiling of patient-derived brain tumors, we identify the downstream metabolism of tryptophan and accumulation of quinolinate (QA) as a metabolic node in glioblastoma and demonstrate its critical role in promoting immune tolerance. QA acts as a metabolic checkpoint in glioblastoma by inducing NMDA receptor activation and Foxo1/PPARγ signaling in macrophages, resulting in a tumor supportive phenotype. Using a genetically-engineered mouse model designed to inhibit production of QA, we identify kynureninase as a promising therapeutic target to revert the potent immune suppressive microenvironment in glioblastoma. These findings offer an opportunity to revisit the biologic consequence of this pathway as it relates to oncogenesis and neurodegenerative disease and a framework for developing immune modulatory agents to further clinical gains in these otherwise incurable diseases.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

