A urine-based ELISA with recombinant non-glycosylated SARS-CoV-2 spike protein for detecting anti-SARS-CoV-2 spike antibodies
- PMID: 36927952
- PMCID: PMC10018619
- DOI: 10.1038/s41598-023-31382-5
A urine-based ELISA with recombinant non-glycosylated SARS-CoV-2 spike protein for detecting anti-SARS-CoV-2 spike antibodies
Abstract
Serological assays have been widely used to detect anti-SARS-CoV-2 antibodies, which are generated from previous exposure to the virus or after vaccination. The presence of anti-SARS-CoV-2 Nucleocapsid antibodies was recently reported in patients´ urine using an in-house urine-based ELISA-platform, allowing a non-invasive way to collect clinical samples and assess immune conversion. In the current study, we evaluated and validated another in-house urine-based ELISA for the detection of anti-SARS-CoV-2 Spike antibodies. Three partial recombinant SARS-CoV-2 Spike proteins comprising the Receptor Binding Domain, expressed in eukaryotic or prokaryotic systems, were tested in an ELISA platform against a panel of over 140 urine and paired serum samples collected from 106 patients confirmed positive for SARS-CoV-2 by qRT-PCR. The key findings from our study were that anti-SARS-CoV-2 Spike antibodies could be detected in urine samples and that the prokaryotic expression of the rSARS-CoV-2 Spike protein was not a barrier to obtain relatively high serology efficiency for the urine-based assay. Thus, use of a urine-based ELISA assay with partial rSARS-CoV-2 Spike proteins, expressed in a prokaryotic system, could be considered as a convenient tool for screening for the presence of anti-SARS-CoV-2 Spike antibodies, and overcome the difficulties arising from sample collection and the need for recombinant proteins produced with eukaryotic expression systems.
© 2023. The Author(s).
Conflict of interest statement
A patent application to the Brazilian National Institute of Industrial Property was issued by UFMG under case number BR 10 2021 014066 6 with FFR, FFB, PFV, JAOS, TARR, CGR, VN, FGF, EAFC and FL as inventors. The authors declare that they have no other competing of interests.
Figures



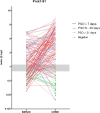
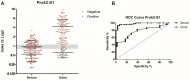
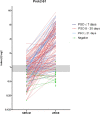
References
-
- WHO. Coronavirus (COVID-19) dashboard. World Health Organization. https://COVID19.who.int/ (2021).
-
- FDA. EUA authorized serology test performance. Food and Drug Administration. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-em... (2021)
-
- WHO. Antigen-detection in the diagnosis of SARS-CoV-2 infection. World Health Organization. https://www.who.int/publications/i/item/antigen-detection-in-the-diagnos... (2021).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

