Rye: genetic ancestry inference at biobank scale
- PMID: 36928108
- PMCID: PMC10164567
- DOI: 10.1093/nar/gkad149
Rye: genetic ancestry inference at biobank scale
Abstract
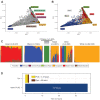
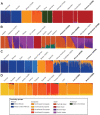
Biobank projects are generating genomic data for many thousands of individuals. Computational methods are needed to handle these massive data sets, including genetic ancestry (GA) inference tools. Current methods for GA inference do not scale to biobank-size genomic datasets. We present Rye-a new algorithm for GA inference at biobank scale. We compared the accuracy and runtime performance of Rye to the widely used RFMix, ADMIXTURE and iAdmix programs and applied it to a dataset of 488221 genome-wide variant samples from the UK Biobank. Rye infers GA based on principal component analysis of genomic variant samples from ancestral reference populations and query individuals. The algorithm's accuracy is powered by Metropolis-Hastings optimization and its speed is provided by non-negative least squares regression. Rye produces highly accurate GA estimates for three-way admixed populations-African, European and Native American-compared to RFMix and ADMIXTURE (${R}^2 = \ 0.998 - 1.00$), and shows 50× runtime improvement compared to ADMIXTURE on the UK Biobank dataset. Rye analysis of UK Biobank samples demonstrates how it can be used to infer GA at both continental and subcontinental levels. We discuss user consideration and options for the use of Rye; the program and its documentation are distributed on the GitHub repository: https://github.com/healthdisparities/rye.
Published by Oxford University Press on behalf of Nucleic Acids Research 2023.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

