A pangenome approach-based loop-mediated isothermal amplification assay for the specific and early detection of Bordetella pertussis
- PMID: 36928221
- PMCID: PMC10018623
- DOI: 10.1038/s41598-023-29773-9
A pangenome approach-based loop-mediated isothermal amplification assay for the specific and early detection of Bordetella pertussis
Abstract
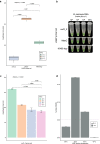
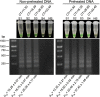
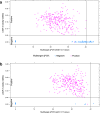
Despite widespread vaccination, Bordetella pertussis continues to cause pertussis infections worldwide, leaving infants at the highest risk of severe illness and death, while people around them are likely the main sources of infection and rapidly spread the disease. Rapid and less complex molecular testing for the specific and timely diagnosis of pertussis remains a challenge that could help to prevent the disease from worsening and prevent its transmission. We aimed to develop and validate a colorimetric loop-mediated isothermal amplification (LAMP) assay using a new target uvrD_2 informed by the pangenome for the specific and early detection of B. pertussis. Compared to that of multitarget quantitative polymerase chain reaction (multitarget qPCR) using a large clinical DNA specimen (n = 600), the diagnostic sensitivity and specificity of the uvrD_2 LAMP assay were 100.0% and 98.6%, respectively, with a 99.7% degree of agreement between the two assays. The novel colorimetric uvrD_2 LAMP assay is highly sensitive and specific for detecting B. pertussis DNA in nasopharyngeal swabs and showed similar diagnostic accuracy to complex and high-cost multitarget qPCR, but it is faster, simpler, and inexpensive, which makes it very helpful for the reliable and timely diagnosis of pertussis in primary health care and resource-limited settings.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- World Health Organization. Pertussis vaccines: WHO position paper. 35(90):433–60 https://www.who.int/publications/i/item/WHO-WER9035 (2015).
-
- Gentile A, et al. Epidemiology of Bordetella pertussis in a children’s hospital. Arch. Argent Pediatr. 2014;112:26–32. - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

