MafB-restricted local monocyte proliferation precedes lung interstitial macrophage differentiation
- PMID: 36928411
- PMCID: PMC10154211
- DOI: 10.1038/s41590-023-01468-3
MafB-restricted local monocyte proliferation precedes lung interstitial macrophage differentiation
Abstract
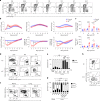
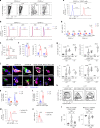
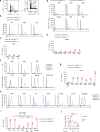
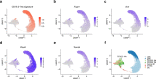
Resident tissue macrophages (RTMs) are differentiated immune cells that populate distinct niches and exert important tissue-supportive functions. RTM maintenance is thought to rely either on differentiation from monocytes or on RTM self-renewal. Here, we used a mouse model of inducible lung interstitial macrophage (IM) niche depletion and refilling to investigate the development of IMs in vivo. Using time-course single-cell RNA-sequencing analyses, bone marrow chimeras and gene targeting, we found that engrafted Ly6C+ classical monocytes proliferated locally in a Csf1 receptor-dependent manner before differentiating into IMs. The transition from monocyte proliferation toward IM subset specification was controlled by the transcription factor MafB, while c-Maf specifically regulated the identity of the CD206+ IM subset. Our data provide evidence that, in the mononuclear phagocyte system, the ability to proliferate is not merely restricted to myeloid progenitor cells and mature RTMs but is also a tightly regulated capability of monocytes developing into RTMs in vivo.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









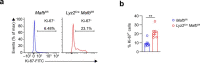

Comment in
-
Macrophage niche availability enables local monocyte proliferation in peripheral tissues.Nat Immunol. 2023 May;24(5):743-745. doi: 10.1038/s41590-023-01482-5. Nat Immunol. 2023. PMID: 36997673 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

