A beta cell subset with enhanced insulin secretion and glucose metabolism is reduced in type 2 diabetes
- PMID: 36928765
- PMCID: PMC10449536
- DOI: 10.1038/s41556-023-01103-1
A beta cell subset with enhanced insulin secretion and glucose metabolism is reduced in type 2 diabetes
Abstract
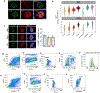
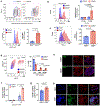
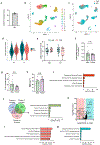
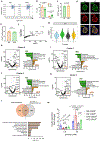
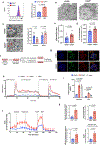
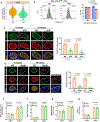
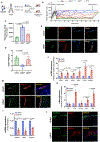
The pancreatic islets are composed of discrete hormone-producing cells that orchestrate systemic glucose homeostasis. Here we identify subsets of beta cells using a single-cell transcriptomic approach. One subset of beta cells marked by high CD63 expression is enriched for the expression of mitochondrial metabolism genes and exhibits higher mitochondrial respiration compared with CD63lo beta cells. Human and murine pseudo-islets derived from CD63hi beta cells demonstrate enhanced glucose-stimulated insulin secretion compared with pseudo-islets from CD63lo beta cells. We show that CD63hi beta cells are diminished in mouse models of and in humans with type 2 diabetes. Finally, transplantation of pseudo-islets generated from CD63hi but not CD63lo beta cells into diabetic mice restores glucose homeostasis. These findings suggest that loss of a specific subset of beta cells may lead to diabetes. Strategies to reconstitute or maintain CD63hi beta cells may represent a potential anti-diabetic therapy.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Declaration of Interests
No competing interests from the authors.
Figures








References
-
- Festa A, Williams K, D'Agostino R Jr., Wagenknecht LE & Haffner SM The natural course of beta-cell function in nondiabetic and diabetic individuals: the Insulin Resistance Atherosclerosis Study. Diabetes 55, 1114–1120 (2006). - PubMed
-
- Ahlqvist E et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6, 361–369 (2018). - PubMed
-
- Wagner R et al. Pathophysiology-based subphenotyping of individuals at elevated risk for type 2 diabetes. Nat Med 27, 49–57 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

