Neoadjuvant chemotherapy plus nivolumab with or without ipilimumab in operable non-small cell lung cancer: the phase 2 platform NEOSTAR trial
- PMID: 36928818
- PMCID: PMC10033402
- DOI: 10.1038/s41591-022-02189-0
Neoadjuvant chemotherapy plus nivolumab with or without ipilimumab in operable non-small cell lung cancer: the phase 2 platform NEOSTAR trial
Abstract
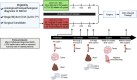
Neoadjuvant ipilimumab + nivolumab (Ipi+Nivo) and nivolumab + chemotherapy (Nivo+CT) induce greater pathologic response rates than CT alone in patients with operable non-small cell lung cancer (NSCLC). The impact of adding ipilimumab to neoadjuvant Nivo+CT is unknown. Here we report the results and correlates of two arms of the phase 2 platform NEOSTAR trial testing neoadjuvant Nivo+CT and Ipi+Nivo+CT with major pathologic response (MPR) as the primary endpoint. MPR rates were 32.1% (7/22, 80% confidence interval (CI) 18.7-43.1%) in the Nivo+CT arm and 50% (11/22, 80% CI 34.6-61.1%) in the Ipi+Nivo+CT arm; the primary endpoint was met in both arms. In patients without known tumor EGFR/ALK alterations, MPR rates were 41.2% (7/17) and 62.5% (10/16) in the Nivo+CT and Ipi+Nivo+CT groups, respectively. No new safety signals were observed in either arm. Single-cell sequencing and multi-platform immune profiling (exploratory endpoints) underscored immune cell populations and phenotypes, including effector memory CD8+ T, B and myeloid cells and markers of tertiary lymphoid structures, that were preferentially increased in the Ipi+Nivo+CT cohort. Baseline fecal microbiota in patients with MPR were enriched with beneficial taxa, such as Akkermansia, and displayed reduced abundance of pro-inflammatory and pathogenic microbes. Neoadjuvant Ipi+Nivo+CT enhances pathologic responses and warrants further study in operable NSCLC. (ClinicalTrials.gov registration: NCT03158129 .).
© 2023. The Author(s).
Conflict of interest statement
T.C. reports speaker fees/honoraria from the Society for Immunotherapy of Cancer, Bristol Myers Squibb, Roche, Medscape, IDEOlogy Health, OncLive, Physicians' Education Resource and PeerView; travel, food and beverage expenses from the Physicians’ Education Resource, Dava Oncology, IDEOlogy Health and Bristol Myers Squibb; advisory role/consulting fees from MedImmune/AstraZeneca, Bristol Myers Squibb, EMD Serono, Merck, Genentech, Arrowhead Pharmaceuticals and Regeneron; and institutional research funding from MedImmune/AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim and EMD Serono. M.C.B.G. has received research funding from Siemens Healthcare. W.N.W. reports consulting or advisory role fees from Clovis Oncology and AstraZeneca; speaker’s fees from Boehringer Ingelheim; honoraria from Roche/Genentech, AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Merck, Bayer, Pfizer and Eli Lilly; and research funding from OSI Pharmaceuticals, Boehringer Ingelheim, Bristol Myers Squibb, Eli Lilly and Merck. M.V.N. reports institutional research funding from Mirati, Novartis, Checkmate, Alaunos/Ziopharm, AstraZeneca, Pfizer and Genentech and consultant/advisory board participation from Mirati, Merck/Merck Sharp & Dohme, Novartis and Genentech. X.L. receives consultant and advisory fees from Eli Lilly, AstraZeneca and EMD Serono and research funding from Eli Lilly, Boehringer Ingelheim and Spectrum Pharmaceuticals. C.M.G. reports fees for advisory committees from AstraZeneca, Bristol Myers Squibb, Jazz Pharmaceuticals and Monte Rosa Therapeutics; research support from AstraZeneca; and speaker’s fees from AstraZeneca and BeiGene. A.S.T. reports personal fees from Bristol Myers Squibb, Eli Lilly, Roche, Novartis, Ariad, EMD Serono, Merck, Seattle Genetics, AstraZeneca, Boehringer Ingelheim, Sellas Life Science and Takeda and grants from Millennium, Polaris, Epizyme and EMD Serono, all outside the submitted work. L.A.B. receives consulting or advisory board fees from Merck Sharp & Dohme, Arrowhead Pharmaceuticals, Chugai Pharmaceutical Company, AstraZeneca, Genentech, BeiGene, AbbVie and Jazz Pharmaceuticals and research funding support from AstraZeneca. M.A. reports research funding to MD Anderson Cancer Center from Genentech, Nektar Therapeutics, Merck, GlaxoSmithKline, Novartis, Jounce Therapeutics, Bristol Myers Squibb, Eli Lilly and Adaptimmune and receives advisory fees from GlaxoSmithKline and Shattuck Labs. B.S.G. reports research funding to MD Anderson Cancer Center from Pfizer, ISA Pharmaceuticals, MedImmune/AstraZeneca and Cue Bio. G.B. receives personal fees and research funding from Amgen, Bayer, Bristol Myers Squibb, Celgene, CytomX Therapeutics, Daiichi Sankyo, Genentech, MedImmune, Merck, Roche and Xcovery; research funding from Adaptimmune, AstraZeneca, Exelixis, GlaxoSmithKline, Immatics, Immunocore, Incyte, Kite Pharma, Macrogenics, Torque, Duality Biologics, Tmunity, Regeneron, Sanofi, BeiGene, Novartis, Verastem and Repertoire Immune Medicines; and personal fees from AbbVie, Adicet, Amgen, Aiad, Clovis Oncology, AstraZeneca, BeiGene, Bristol Myers Squibb, Celgene, Genentech, Genzyme, Gilead, Instil Bio, Intervenn Biosciences, Eli Lilly, Merck, Novartis, Roche, Onconova Therapeutics, Regeneron, Sanofi, Tyme Oncology, Xcovery, Virogin Biotech, Johnson & Johnson/Janssen and Maverick Therapeutics. J.Z. receives research funding from Merck, Johnson & Johnson and Novartis and consulting fees/speaker fees/honoraria from Bristol Myers Squibb, AstraZeneca, Roche, GenePlus, Innovent and Hengrui, all outside the submitted work. W.L.H. receives research funding from Johnson & Johnson. M.A. reports consulting fees from AstraZeneca. C.H. reports speaker’s fees from the Society for Immunotherapy of Cancer; research funding to the institution from Iovance, Sanofi, BTG and Dragonfly; stock options for Briacell; advisory board membership for Briacell and the Mesothelioma Applied Research Foundation; and personal fees from Nanobiotix. I.I.W. reports honoraria from Genentech/Roche, Bayer, Bristol Myers Squibb, AstraZeneca/Medimmune, Pfizer, HTG Molecular, Asuragen, Merck, GlaxoSmithKline, Guardant Health, Platform Health, Daiichi, Merck, Flame, Oncocyte and Merck Sharp & Dohme and research support from Genentech, Oncoplex, HTG Molecular, DepArray, Merck, Bristol Myers Squibb, Medimmune, Adaptive, Adaptimmune, EMD Serono, Pfizer, Takeda, Amgen, Karus, Johnson & Johnson, Bayer, Iovance, 4D Pharma, Novartis and Akoya. S.G.S. reports speaker, travel and lodging expenses from the Egyptian Society of Surgical Oncology/Best of SSO Cairo and the West Hawaii Cancer Symposium; review panel participation and travel and lodging expenses from the Peter MacCallum Cancer Centre; and unpaid advisory board participation for Ethicon. D.L.G. reports honoraria for scientific advisory boards from AstraZeneca, Sanofi, Alethia Biotherapeutics, Menarini, Eli Lilly, 4D Pharma and Onconova and research support from Janssen, Takeda, Astellas, Ribon Therapeutics, NGM Biopharmaceuticals, Boehringer Ingelheim, Mirati Therapeutics and AstraZeneca. J.A.W. is an inventor on a US patent application (PCT/US17/53.717) submitted by The University of Texas MD Anderson Cancer Center that covers methods to enhance immune checkpoint blockade responses by modulating the microbiome. J.A.W. also reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, Physician Education Resource, MedImmune, Exelixis and Bristol Myers Squibb; serves as a consultant/advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline, Bristol Myers Squibb, Merck, Biothera Pharmaceuticals and Microbiome DX; and receives research support from GlaxoSmithKline, Roche/Genentech, Bristol Myers Squibb and Novartis. J.P.A. reports consulting, stock ownership or advisory board membership from Achelois, Adaptive Biotechnologies, Apricity, BioAtla, BioNTech, Candel Therapeutics, Codiak Biosciences, Dragonfly Therapeutics, Earli, Enable Medicine, Hummingbird, ImaginAb, Lava Therapeutics, Lytix Biopharma, Marker Therapeutics, PBM Capital, Phenomic AI, Polaris Pharma, Time Bioventures, Trained Therapeutix Discovery, Two Bear Capital and InterVenn Biosciences. P.S. reports consulting, stock ownership or advisory board membership from Achelois, Adaptive Biotechnologies, Affini-T, Apricity, Asher Bio, BioAtla, BioNTech, Candel Therapeutics, Carisma, Catalio, Codiak Biosciences, C-Reveal Therapeutics, Dragonfly Therapeutics, Earli, Enable Medicine, Glympse, Henlius/Hengenix, Hummingbird, ImaginAb, Infinity Pharma, InterVenn Biosciences, JSL Health, Lava Therapeutics, Lytix Biopharma, Marker Therapeutics, Oncolytics, PBM Capital, Phenomic AI, Polaris Pharma, Sporos, Time Bioventures, Trained Therapeutix Discovery, Two Bear Capital and Xilis. H.K. receives funding to MD Anderson Cancer Center from Johnson & Johnson. J.V.H. reports fees for advisory committees/consulting from AstraZeneca, EMD Serono, Boehringer Ingelheim, Catalyst, Genentech, GlaxoSmithKline, Hengrui Therapeutics, Eli Lilly, Spectrum, Sanofi, Takeda, Mirati Therapeutics, Bristol Myers Squibb, BrightPath Biotherapeutics, Janssen Global Services, Nexus Health Systems, Pneuma Respiratory, Kairos Venture Investments, Roche, Leads Biolabs, RefleXion and Chugai Pharmaceutical Company; research support from AstraZeneca, Bristol Myers Squibb, Spectrum and Takeda; and royalties and licensing fees from Spectrum. B.S. reports consulting and speaker fees from PeerView, AstraZeneca and Medscape and institutional research funding from Bristol Myers Squibb. The other authors report no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

