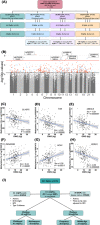
Gestational weight gain in pregnant women with obesity is associated with cord blood DNA methylation, which partially mediates offspring anthropometrics
- PMID: 36929108
- PMCID: PMC10019770
- DOI: 10.1002/ctm2.1215
Gestational weight gain in pregnant women with obesity is associated with cord blood DNA methylation, which partially mediates offspring anthropometrics
Keywords: birthweight; epigenetics; fetal programming; lean mass.
Figures

References
-
- Rasmussen KM, Yaktine AL, eds. Weight Gain During Pregnancy: Reexamining the Guidelines . National Academies Press (US); 2009. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
