2022 updates to the Rat Genome Database: a Findable, Accessible, Interoperable, and Reusable (FAIR) resource
- PMID: 36930729
- PMCID: PMC10474928
- DOI: 10.1093/genetics/iyad042
2022 updates to the Rat Genome Database: a Findable, Accessible, Interoperable, and Reusable (FAIR) resource
Abstract
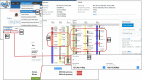
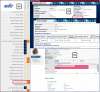
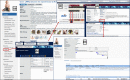
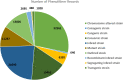
The Rat Genome Database (RGD, https://rgd.mcw.edu) has evolved from simply a resource for rat genetic markers, maps, and genes, by adding multiple genomic data types and extensive disease and phenotype annotations and developing tools to effectively mine, analyze, and visualize the available data, to empower investigators in their hypothesis-driven research. Leveraging its robust and flexible infrastructure, RGD has added data for human and eight other model organisms (mouse, 13-lined ground squirrel, chinchilla, naked mole-rat, dog, pig, African green monkey/vervet, and bonobo) besides rat to enhance its translational aspect. This article presents an overview of the database with the most recent additions to RGD's genome, variant, and quantitative phenotype data. We also briefly introduce Virtual Comparative Map (VCMap), an updated tool that explores synteny between species as an improvement to RGD's suite of tools, followed by a discussion regarding the refinements to the existing PhenoMiner tool that assists researchers in finding and comparing quantitative data across rat strains. Collectively, RGD focuses on providing a continuously improving, consistent, and high-quality data resource for researchers while advancing data reproducibility and fulfilling Findable, Accessible, Interoperable, and Reusable (FAIR) data principles.
Keywords: Rattus norvegicus; FAIR data; Rat Genome Database; comparative genome; genomics; quantitative phenotype; rat genetics; rat strain.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Genetics Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

