Restoration of Motor Function through Delayed Intraspinal Delivery of Human IL-10-Encoding Nucleoside-Modified mRNA after Spinal Cord Injury
- PMID: 36930811
- PMCID: PMC10013810
- DOI: 10.34133/research.0056
Restoration of Motor Function through Delayed Intraspinal Delivery of Human IL-10-Encoding Nucleoside-Modified mRNA after Spinal Cord Injury
Abstract
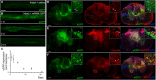
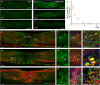
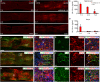
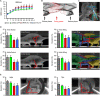
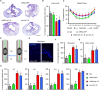
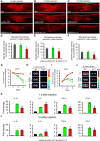
Efficient in vivo delivery of anti-inflammatory proteins to modulate the microenvironment of an injured spinal cord and promote neuroprotection and functional recovery is a great challenge. Nucleoside-modified messenger RNA (mRNA) has become a promising new modality that can be utilized for the safe and efficient delivery of therapeutic proteins. Here, we used lipid nanoparticle (LNP)-encapsulated human interleukin-10 (hIL-10)-encoding nucleoside-modified mRNA to induce neuroprotection and functional recovery following rat spinal cord contusion injury. Intralesional administration of hIL-10 mRNA-LNP to rats led to a remarkable reduction of the microglia/macrophage reaction in the injured spinal segment and induced significant functional recovery compared to controls. Furthermore, hIL-10 mRNA treatment induced increased expression in tissue inhibitor of matrix metalloproteinase 1 and ciliary neurotrophic factor levels in the affected spinal segment indicating a time-delayed secondary effect of IL-10 5 d after injection. Our results suggest that treatment with nucleoside-modified mRNAs encoding neuroprotective factors is an effective strategy for spinal cord injury repair.
Copyright © 2023 László Gál et al.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources

