Real-World Validation of Molecular International Prognostic Scoring System for Myelodysplastic Syndromes
- PMID: 36930857
- PMCID: PMC10414702
- DOI: 10.1200/JCO.22.01784
Real-World Validation of Molecular International Prognostic Scoring System for Myelodysplastic Syndromes
Abstract
Purpose: Myelodysplastic syndromes (MDS) are heterogeneous myeloid neoplasms in which a risk-adapted treatment strategy is needed. Recently, a new clinical-molecular prognostic model, the Molecular International Prognostic Scoring System (IPSS-M) was proposed to improve the prediction of clinical outcome of the currently available tool (Revised International Prognostic Scoring System [IPSS-R]). We aimed to provide an extensive validation of IPSS-M.
Methods: A total of 2,876 patients with primary MDS from the GenoMed4All consortium were retrospectively analyzed.
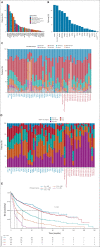
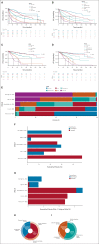
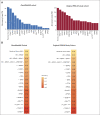
Results: IPSS-M improved prognostic discrimination across all clinical end points with respect to IPSS-R (concordance was 0.81 v 0.74 for overall survival and 0.89 v 0.76 for leukemia-free survival, respectively). This was true even in those patients without detectable gene mutations. Compared with the IPSS-R based stratification, the IPSS-M risk group changed in 46% of patients (23.6% and 22.4% of subjects were upstaged and downstaged, respectively).In patients treated with hematopoietic stem cell transplantation (HSCT), IPSS-M significantly improved the prediction of the risk of disease relapse and the probability of post-transplantation survival versus IPSS-R (concordance was 0.76 v 0.60 for overall survival and 0.89 v 0.70 for probability of relapse, respectively). In high-risk patients treated with hypomethylating agents (HMA), IPSS-M failed to stratify individual probability of response; response duration and probability of survival were inversely related to IPSS-M risk.Finally, we tested the accuracy in predicting IPSS-M when molecular information was missed and we defined a minimum set of 15 relevant genes associated with high performance of the score.
Conclusion: IPSS-M improves MDS prognostication and might result in a more effective selection of candidates to HSCT. Additional factors other than gene mutations can be involved in determining HMA sensitivity. The definition of a minimum set of relevant genes may facilitate the clinical implementation of the score.
Trial registration: ClinicalTrials.gov NCT04889729.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
No other potential conflicts of interest were reported.
Figures


References
-
- Della Porta MG, Tuechler H, Malcovati L, et al. : Validation of WHO classification-based Prognostic Scoring System (WPSS) for myelodysplastic syndromes and comparison with the revised International Prognostic Scoring System (IPSS-R). A study of the International Working Group for Prognosis in Myelodysplasia (IWG-PM). Leukemia 29:1502-1513, 2015 - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

