Synthetic microbial communities (SynComs) of the human gut: design, assembly, and applications
- PMID: 36931888
- PMCID: PMC10062696
- DOI: 10.1093/femsre/fuad012
Synthetic microbial communities (SynComs) of the human gut: design, assembly, and applications
Abstract
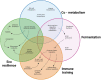
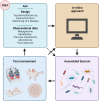
The human gut harbors native microbial communities, forming a highly complex ecosystem. Synthetic microbial communities (SynComs) of the human gut are an assembly of microorganisms isolated from human mucosa or fecal samples. In recent decades, the ever-expanding culturing capacity and affordable sequencing, together with advanced computational modeling, started a ''golden age'' for harnessing the beneficial potential of SynComs to fight gastrointestinal disorders, such as infections and chronic inflammatory bowel diseases. As simplified and completely defined microbiota, SynComs offer a promising reductionist approach to understanding the multispecies and multikingdom interactions in the microbe-host-immune axis. However, there are still many challenges to overcome before we can precisely construct SynComs of designed function and efficacy that allow the translation of scientific findings to patients' treatments. Here, we discussed the strategies used to design, assemble, and test a SynCom, and address the significant challenges, which are of microbiological, engineering, and translational nature, that stand in the way of using SynComs as live bacterial therapeutics.
Keywords: human gut microbiome; live bacterial therapeutic; rational design; synthetic microbial consortium.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Synthetic Communities of Gut Microbes for Basic Research and Translational Approaches in Animal Health and Nutrition.Annu Rev Anim Biosci. 2024 Feb 15;12:283-300. doi: 10.1146/annurev-animal-021022-025552. Epub 2023 Nov 14. Annu Rev Anim Biosci. 2024. PMID: 37963399 Review.
-
Building microbial synthetic communities: get inspired by the design of synthetic plant communities.New Phytol. 2025 Apr;246(2):402-405. doi: 10.1111/nph.70011. Epub 2025 Feb 17. New Phytol. 2025. PMID: 39957629 Free PMC article.
-
Evaluation of ready-to-use freezer stocks of a synthetic microbial community for maize root colonization.Microbiol Spectr. 2024 Jan 11;12(1):e0240123. doi: 10.1128/spectrum.02401-23. Epub 2023 Dec 12. Microbiol Spectr. 2024. PMID: 38084978 Free PMC article.
-
Integrating ecological and evolutionary frameworks for SynCom success.New Phytol. 2025 Jun;246(5):1922-1933. doi: 10.1111/nph.70112. Epub 2025 Apr 3. New Phytol. 2025. PMID: 40177999
-
Starting with screening strains to construct synthetic microbial communities (SynComs) for traditional food fermentation.Food Res Int. 2024 Aug;190:114557. doi: 10.1016/j.foodres.2024.114557. Epub 2024 May 27. Food Res Int. 2024. PMID: 38945561 Review.
Cited by
-
Microbiome modeling: a beginner's guide.Front Microbiol. 2024 Jun 19;15:1368377. doi: 10.3389/fmicb.2024.1368377. eCollection 2024. Front Microbiol. 2024. PMID: 38962127 Free PMC article. Review.
-
Strategies for tailoring functional microbial synthetic communities.ISME J. 2024 Jan 8;18(1):wrae049. doi: 10.1093/ismejo/wrae049. ISME J. 2024. PMID: 38537571 Free PMC article. Review.
-
The Respiratory Tract Microbiome and Human Health.Microb Biotechnol. 2025 May;18(5):e70147. doi: 10.1111/1751-7915.70147. Microb Biotechnol. 2025. PMID: 40293161 Free PMC article. Review.
-
Adaptive and metabolic convergence in rhizosphere and gut microbiomes.Microbiome. 2025 Jul 26;13(1):173. doi: 10.1186/s40168-025-02179-7. Microbiome. 2025. PMID: 40713710 Free PMC article. Review.
-
Present and future of microbiome-targeting therapeutics.J Clin Invest. 2025 Jun 2;135(11):e184323. doi: 10.1172/JCI184323. eCollection 2025 Jun 2. J Clin Invest. 2025. PMID: 40454480 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

