DIAPH1 mediates progression of atherosclerosis and regulates hepatic lipid metabolism in mice
- PMID: 36932214
- PMCID: PMC10023694
- DOI: 10.1038/s42003-023-04643-2
DIAPH1 mediates progression of atherosclerosis and regulates hepatic lipid metabolism in mice
Abstract
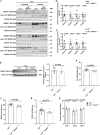
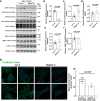
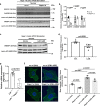
Atherosclerosis evolves through dysregulated lipid metabolism interwoven with exaggerated inflammation. Previous work implicating the receptor for advanced glycation end products (RAGE) in atherosclerosis prompted us to explore if Diaphanous 1 (DIAPH1), which binds to the RAGE cytoplasmic domain and is important for RAGE signaling, contributes to these processes. We intercrossed atherosclerosis-prone Ldlr-/- mice with mice devoid of Diaph1 and fed them Western diet for 16 weeks. Compared to male Ldlr-/- mice, male Ldlr-/- Diaph1-/- mice displayed significantly less atherosclerosis, in parallel with lower plasma concentrations of cholesterol and triglycerides. Female Ldlr-/- Diaph1-/- mice displayed significantly less atherosclerosis compared to Ldlr-/- mice and demonstrated lower plasma concentrations of cholesterol, but not plasma triglycerides. Deletion of Diaph1 attenuated expression of genes regulating hepatic lipid metabolism, Acaca, Acacb, Gpat2, Lpin1, Lpin2 and Fasn, without effect on mRNA expression of upstream transcription factors Srebf1, Srebf2 or Mxlipl in male mice. We traced DIAPH1-dependent mechanisms to nuclear translocation of SREBP1 in a manner independent of carbohydrate- or insulin-regulated cues but, at least in part, through the actin cytoskeleton. This work unveils new regulators of atherosclerosis and lipid metabolism through DIAPH1.
© 2023. The Author(s).
Conflict of interest statement
R.R., A.S., M.B.M., and A.M.S. have patents and patent applications through NYU Grossman School of Medicine that have been submitted/published that are indirectly related to the work detailed in this manuscript.
Figures
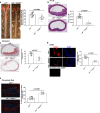
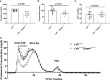
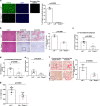
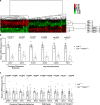

References
-
- Heron, M. Deaths: Leading Causes for 2017. Natl. Vital Stat. Rep.68, 1–77 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

