Investigation of targets and anticancer mechanisms of covalently acting natural products by functional proteomics
- PMID: 36932232
- PMCID: PMC10374574
- DOI: 10.1038/s41401-023-01072-z
Investigation of targets and anticancer mechanisms of covalently acting natural products by functional proteomics
Abstract
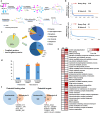
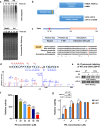
Eriocalyxin B (EB), 17-hydroxy-jolkinolide B (HJB), parthenolide (PN), xanthatin (XT) and andrographolide (AG) are terpenoid natural products with a variety of promising antitumor activities, which commonly bear electrophilic groups (α,β-unsaturated carbonyl groups and/or epoxides) capable of covalently modifying protein cysteine residues. However, their direct targets and underlying molecular mechanisms are still largely unclear, which limits the development of these compounds. In this study, we integrated activity-based protein profiling (ABPP) and quantitative proteomics approach to systematically characterize the covalent targets of these natural products and their involved cellular pathways. We first demonstrated the anti-proliferation activities of these five compounds in triple-negative breast cancer cell MDA-MB-231. Tandem mass tag (TMT)-based quantitative proteomics showed all five compounds commonly affected the ubiquitin mediated proteolysis pathways. ABPP platform identified the preferentially modified targets of EB and PN, two natural products with high anti-proliferation activity. Biochemical experiments showed that PN inhibited the cell proliferation through targeting ubiquitin carboxyl-terminal hydrolase 10 (USP10). Together, this study uncovered the covalently modified targets of these natural products and potential molecular mechanisms of their antitumor activities.
Keywords: ABPP; eriocalyxin B; natural products; parthenolide; proteomics.
© 2023. The Author(s), under exclusive licence to Shanghai Institute of Materia Medica, Chinese Academy of Sciences and Chinese Pharmacological Society.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Target Identification of Bioactive Covalently Acting Natural Products.Curr Top Microbiol Immunol. 2019;420:351-374. doi: 10.1007/82_2018_121. Curr Top Microbiol Immunol. 2019. PMID: 30105423 Review.
-
Comprehensive proteomics and sialiomics of the anti-proliferative activity of safranal on triple negative MDA-MB-231 breast cancer cell lines.J Proteomics. 2022 May 15;259:104539. doi: 10.1016/j.jprot.2022.104539. Epub 2022 Feb 28. J Proteomics. 2022. PMID: 35240313
-
Parthenolide Covalently Targets and Inhibits Focal Adhesion Kinase in Breast Cancer Cells.Cell Chem Biol. 2019 Jul 18;26(7):1027-1035.e22. doi: 10.1016/j.chembiol.2019.03.016. Epub 2019 May 9. Cell Chem Biol. 2019. PMID: 31080076 Free PMC article.
-
Harnessing the anti-cancer natural product nimbolide for targeted protein degradation.Nat Chem Biol. 2019 Jul;15(7):747-755. doi: 10.1038/s41589-019-0304-8. Epub 2019 Jun 17. Nat Chem Biol. 2019. PMID: 31209351 Free PMC article.
-
Covalent Modification of Proteins by Plant-Derived Natural Products: Proteomic Approaches and Biological Impacts.Proteomics. 2021 Feb;21(3-4):e1900386. doi: 10.1002/pmic.201900386. Epub 2020 Dec 31. Proteomics. 2021. PMID: 32949481 Free PMC article. Review.
Cited by
-
Development of target-based cell membrane affinity ultrafiltration technology for a simplified approach to discovering potential bioactive compounds in natural products.Anal Bioanal Chem. 2024 Mar;416(7):1647-1655. doi: 10.1007/s00216-024-05166-3. Epub 2024 Feb 2. Anal Bioanal Chem. 2024. PMID: 38305859
-
Isotoosendanin inhibits triple-negative breast cancer metastasis by reducing mitochondrial fission and lamellipodia formation regulated by the Smad2/3-GOT2-MYH9 signaling axis.Acta Pharmacol Sin. 2024 Dec;45(12):2672-2683. doi: 10.1038/s41401-024-01335-3. Epub 2024 Jul 15. Acta Pharmacol Sin. 2024. PMID: 39009651
-
17-hydroxy-jolkinolide B potentiated CTLA4ab therapy through targeting tumor suppression and immune activation by downregulating PD-L1 expression in lung adenocarcinoma.J Thorac Dis. 2024 Nov 30;16(11):7526-7538. doi: 10.21037/jtd-24-781. Epub 2024 Nov 29. J Thorac Dis. 2024. PMID: 39678884 Free PMC article.
-
Characterization of endogenous SUMOylation sites by click chemistry-based proteomics.Anal Bioanal Chem. 2025 Aug;417(19):4419-4433. doi: 10.1007/s00216-025-05957-2. Epub 2025 Jun 16. Anal Bioanal Chem. 2025. PMID: 40522366
-
Quantitation of global histone post-translational modifications reveal anti-inflammatory epigenetic mechanisms of liquiritigenin based on the optimized super-SILAC strategy.Front Cell Dev Biol. 2025 Mar 27;13:1566567. doi: 10.3389/fcell.2025.1566567. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40213392 Free PMC article.
References
-
- Nomura DK, Maimone TJ. Target identification of bioactive covalently acting natural products. Curr Top Microbiol Immunol. 2019;420:351–74.. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

