Genome-wide identification and characterization of members of the LEA gene family in Panax notoginseng and their transcriptional responses to dehydration of recalcitrant seeds
- PMID: 36932328
- PMCID: PMC10024439
- DOI: 10.1186/s12864-023-09229-0
Genome-wide identification and characterization of members of the LEA gene family in Panax notoginseng and their transcriptional responses to dehydration of recalcitrant seeds
Abstract
Background: Late embryogenesis abundant (LEA) proteins play an important role in dehydration process of seed maturation. The seeds of Panax notoginseng (Burkill) F. H. Chen are typically characterized with the recalcitrance and are highly sensitive to dehydration. However, it is not very well known about the role of LEA proteins in response to dehydration stress in P. notoginseng seeds. We will perform a genome-wide analysis of the LEA gene family and their transcriptional responses to dehydration stress in recalcitrant P. notoginseng seeds.
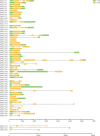
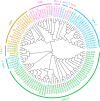
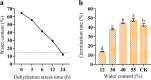
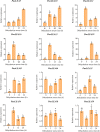
Results: In this study, 61 LEA genes were identified from the P. notoginseng genome, and they were renamed as PnoLEA. The PnoLEA genes were classified into seven subfamilies based on the phylogenetic relationships, gene structure and conserved domains. The PnoLEA genes family showed relatively few introns and was highly conserved. Unexpectedly, the LEA_6 subfamily was not found, and the LEA_2 subfamily contained 46 (75.4%) members. Within 19 pairs of fragment duplication events, among them 17 pairs were LEA_2 subfamily. In addition, the expression of the PnoLEA genes was obviously induced under dehydration stress, but the germination rate of P. notoginseng seeds decreased as the dehydration time prolonged.
Conclusions: We found that the lack of the LEA_6 subfamily, the expansion of the LEA_2 subfamily and low transcriptional levels of most PnoLEA genes might be implicated in the recalcitrant formation of P. notoginseng seeds. LEA proteins are essential in the response to dehydration stress in recalcitrant seeds, but the protective effect of LEA protein is not efficient. These results could improve our understanding of the function of LEA proteins in the response of dehydration stress and their contributions to the formation of seed recalcitrance.
Keywords: Dehydration stress; Expression patterns; LEA; Panax notoginseng; Recalcitrant seeds.
© 2023. The Author(s).
Conflict of interest statement
The authors report no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Exogenous gibberellic acid shortening after-ripening process and promoting seed germination in a medicinal plant Panax notoginseng.BMC Plant Biol. 2023 Feb 1;23(1):67. doi: 10.1186/s12870-023-04084-3. BMC Plant Biol. 2023. PMID: 36721119 Free PMC article.
-
iTRAQ and RNA-seq analyses provide an insight into mechanisms of recalcitrance in a medicinal plant Panax notoginseng seeds during the after-ripening process.Funct Plant Biol. 2021 Dec;49(1):68-88. doi: 10.1071/FP21197. Funct Plant Biol. 2021. PMID: 34822750
-
Genome-wide identification and expression analyses of the LEA protein gene family in tea plant reveal their involvement in seed development and abiotic stress responses.Sci Rep. 2019 Oct 1;9(1):14123. doi: 10.1038/s41598-019-50645-8. Sci Rep. 2019. PMID: 31575979 Free PMC article.
-
Genome-wide identification and expression analysis of MYB gene family under nitrogen stress in Panax notoginseng.Protoplasma. 2023 Jan;260(1):189-205. doi: 10.1007/s00709-022-01770-1. Epub 2022 May 7. Protoplasma. 2023. PMID: 35524823
-
Plant Group II LEA Proteins: Intrinsically Disordered Structure for Multiple Functions in Response to Environmental Stresses.Biomolecules. 2021 Nov 9;11(11):1662. doi: 10.3390/biom11111662. Biomolecules. 2021. PMID: 34827660 Free PMC article. Review.
Cited by
-
Comparative proteomic analysis of desiccation responses in recalcitrant Quercus acutissima seeds.Plant Mol Biol. 2025 May 26;115(3):68. doi: 10.1007/s11103-025-01596-4. Plant Mol Biol. 2025. PMID: 40418286
-
Glutathione S-transferase in mediating adaptive responses of oats (Avena sativa) to osmotic and cadmium stress: genome-wide analysis.BMC Plant Biol. 2025 Apr 25;25(1):538. doi: 10.1186/s12870-025-06559-x. BMC Plant Biol. 2025. PMID: 40281415 Free PMC article.
-
The Late Embryogenesis Abundant Proteins in Soybean: Identification, Expression Analysis, and the Roles of GmLEA4_19 in Drought Stress.Int J Mol Sci. 2023 Oct 2;24(19):14834. doi: 10.3390/ijms241914834. Int J Mol Sci. 2023. PMID: 37834282 Free PMC article.
References
-
- Li X, Cao J. Late Embryogenesis Abundant (LEA) gene family in maize: identification, evolution, and expression profiles. Plant Mol Biol Report. 2016;34(1):15–28. doi: 10.1007/s11105-015-0901-y. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

