HIV-1 Tat amino acid residues that influence Tat-TAR binding affinity: a scoping review
- PMID: 36932337
- PMCID: PMC10020771
- DOI: 10.1186/s12879-023-08123-0
HIV-1 Tat amino acid residues that influence Tat-TAR binding affinity: a scoping review
Abstract
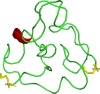
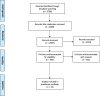
HIV-1 remains a global health concern and to date, nearly 38 million people are living with HIV. The complexity of HIV-1 pathogenesis and its subsequent prevalence is influenced by several factors including the HIV-1 subtype. HIV-1 subtype variation extends to sequence variation in the amino acids of the HIV-1 viral proteins. Of particular interest is the transactivation of transcription (Tat) protein due to its key function in viral transcription. The Tat protein predominantly functions by binding to the transactivation response (TAR) RNA element to activate HIV-1 transcriptional elongation. Subtype-specific Tat protein sequence variation influences Tat-TAR binding affinity. Despite several studies investigating Tat-TAR binding, it is not clear which regions of the Tat protein and/or individual Tat amino acid residues may contribute to TAR binding affinity. We, therefore, conducted a scoping review on studies investigating Tat-TAR binding. We aimed to synthesize the published data to determine (1) the regions of the Tat protein that may be involved in TAR binding, (2) key Tat amino acids involved in TAR binding and (3) if Tat subtype-specific variation influences TAR binding. A total of thirteen studies met our inclusion criteria and the key findings were that (1) both N-terminal and C-terminal amino acids outside the basic domain (47-59) may be important in increasing Tat-TAR binding affinity, (2) substitution of the amino acids Lysine and Arginine (47-59) resulted in a reduction in binding affinity to TAR, and (3) none of the included studies have investigated Tat subtype-specific substitutions and therefore no commentary could be made regarding which subtype may have a higher Tat-TAR binding affinity. Future studies investigating Tat-TAR binding should therefore use full-length Tat proteins and compare subtype-specific variations. Studies of such a nature may help explain why we see differential pathogenesis and prevalence when comparing HIV-1 subtypes.
Keywords: Molecular binding; Subtype variation; Tat polymorphism; Transactivation of transcription; Transactivation response RNA element.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Impact of subtype C-specific amino acid variants on HIV-1 Tat-TAR interaction: insights from molecular modelling and dynamics.Virol J. 2024 Jun 25;21(1):144. doi: 10.1186/s12985-024-02419-6. Virol J. 2024. PMID: 38918875 Free PMC article.
-
CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA.Mol Cell Biol. 2000 Sep;20(18):6958-69. doi: 10.1128/MCB.20.18.6958-6969.2000. Mol Cell Biol. 2000. PMID: 10958691 Free PMC article.
-
Distinct transcriptional pathways of TAR-dependent and TAR-independent human immunodeficiency virus type-1 transactivation by Tat.Virology. 1997 Aug 18;235(1):48-64. doi: 10.1006/viro.1997.8672. Virology. 1997. PMID: 9300036
-
Face-time with TAR: Portraits of an HIV-1 RNA with diverse modes of effector recognition relevant for drug discovery.J Biol Chem. 2019 Jun 14;294(24):9326-9341. doi: 10.1074/jbc.REV119.006860. Epub 2019 May 12. J Biol Chem. 2019. PMID: 31080171 Free PMC article. Review.
-
Genetic variation and function of the HIV-1 Tat protein.Med Microbiol Immunol. 2019 Apr;208(2):131-169. doi: 10.1007/s00430-019-00583-z. Epub 2019 Mar 5. Med Microbiol Immunol. 2019. PMID: 30834965 Free PMC article. Review.
Cited by
-
FUBP3 enhances HIV-1 transcriptional activity and regulates immune response pathways in T cells.Mol Ther Nucleic Acids. 2025 Mar 25;36(2):102525. doi: 10.1016/j.omtn.2025.102525. eCollection 2025 Jun 10. Mol Ther Nucleic Acids. 2025. PMID: 40248217 Free PMC article.
-
Molecular insights into the interaction between a disordered protein and a folded RNA.bioRxiv [Preprint]. 2024 Jun 12:2024.06.12.598678. doi: 10.1101/2024.06.12.598678. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Dec 3;121(49):e2409139121. doi: 10.1073/pnas.2409139121. PMID: 38915483 Free PMC article. Updated. Preprint.
-
A pilot investigation of the association between HIV-1 Vpr amino acid sequence diversity and the tryptophan-kynurenine pathway as a potential mechanism for neurocognitive impairment.Virol J. 2024 Feb 23;21(1):47. doi: 10.1186/s12985-024-02313-1. Virol J. 2024. PMID: 38395987 Free PMC article.
-
Solid Phase Synthesis and TAR RNA-Binding Activity of Nucleopeptides Containing Nucleobases Linked to the Side Chains via 1,4-Linked-1,2,3-triazole.Biomedicines. 2024 Mar 3;12(3):570. doi: 10.3390/biomedicines12030570. Biomedicines. 2024. PMID: 38540183 Free PMC article.
-
Variation in HIV-1 Tat and Vpr protein amino acid sequences and its association with vascular health measures in a South African cohort: an exploratory study.Virol J. 2025 Aug 4;22(1):266. doi: 10.1186/s12985-025-02891-8. Virol J. 2025. PMID: 40759960 Free PMC article.
References
-
- Barré-Sinoussi F, Chermann J-C, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vézinet-Brun F, Rouzioux C. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220(4599):868–871. doi: 10.1126/science.6189183. - DOI - PubMed
-
- UNAIDS. Danger: UNAIDS global AIDS update 2022. In: 2022.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

