The function of plant PR1 and other members of the CAP protein superfamily in plant-pathogen interactions
- PMID: 36932700
- PMCID: PMC10189770
- DOI: 10.1111/mpp.13320
The function of plant PR1 and other members of the CAP protein superfamily in plant-pathogen interactions
Abstract
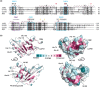
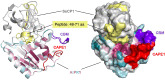
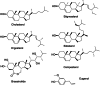
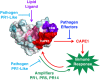
The pathogenesis-related (PR) proteins of plants have originally been identified as proteins that are strongly induced upon biotic and abiotic stress. These proteins fall into 17 distinct classes (PR1-PR17). The mode of action of most of these PR proteins has been well characterized, except for PR1, which belongs to a widespread superfamily of proteins that share a common CAP domain. Proteins of this family are not only expressed in plants but also in humans and in many different pathogens, including phytopathogenic nematodes and fungi. These proteins are associated with a diverse range of physiological functions. However, their precise mode of action has remained elusive. The importance of these proteins in immune defence is illustrated by the fact that PR1 overexpression in plants results in increased resistance against pathogens. However, PR1-like CAP proteins are also produced by pathogens and deletion of these genes results in reduced virulence, suggesting that CAP proteins can exert both defensive and offensive functions. Recent progress has revealed that plant PR1 is proteolytically cleaved to release a C-terminal CAPE1 peptide, which is sufficient to activate an immune response. The release of this signalling peptide is blocked by pathogenic effectors to evade immune defence. Moreover, plant PR1 forms complexes with other PR family members, including PR5, also known as thaumatin, and PR14, a lipid transfer protein, to enhance the host's immune response. Here, we discuss possible functions of PR1 proteins and their interactors, particularly in light of the fact that these proteins can bind lipids, which have important immune signalling functions.
Keywords: effector proteins; fungal CAPs; nematode VALs/VAPs; pathogen virulence; plant PR1; plant immunity; sperm-coating proteins (SCPs).
© 2023 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures
References
-
- Abraham, A. & Chandler, D.E. (2017) Tracing the evolutionary history of the CAP superfamily of proteins using amino acid sequence homology and conservation of splice sites. Journal of Molecular Evolution, 85, 137–157. - PubMed
-
- Alexander, D. , Goodman, R.M. , Gut‐Rella, M. , Glascock, C. , Weymann, K. , Friedrich, L. et al. (1993) Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis‐related protein 1a. Proceedings of the National Academy of Sciences of the United States of America, 90, 7327–7331. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

