Guadecitabine increases response to combined anti-CTLA-4 and anti-PD-1 treatment in mouse melanoma in vivo by controlling T-cells, myeloid derived suppressor and NK cells
- PMID: 36934257
- PMCID: PMC10024396
- DOI: 10.1186/s13046-023-02628-x
Guadecitabine increases response to combined anti-CTLA-4 and anti-PD-1 treatment in mouse melanoma in vivo by controlling T-cells, myeloid derived suppressor and NK cells
Abstract
Background: The combination of Programmed Cell Death 1 (PD-1) and Cytotoxic T-Lymphocyte Antigen 4 (CTLA-4) blockade has dramatically improved the overall survival rate for malignant melanoma. Immune checkpoint blockers (ICBs) limit the tumor's immune escape yet only for approximately a third of all tumors and, in most cases, for a limited amount of time. Several approaches to overcome resistance to ICBs are being investigated among which the addition of epigenetic drugs that are expected to act on both immune and tumor cells. Guadecitabine, a dinucleotide prodrug of a decitabine linked via phosphodiester bond to a guanosine, showed promising results in the phase-1 clinical trial, NIBIT-M4 (NCT02608437).
Methods: We used the syngeneic B16F10 murine melanoma model to study the effects of immune checkpoint blocking antibodies against CTLA-4 and PD-1 in combination, with and without the addition of Guadecitabine. We comprehensively characterized the tumor's and the host's responses under different treatments by flow cytometry, multiplex immunofluorescence and methylation analysis.
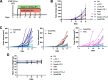
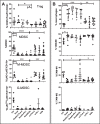
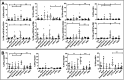
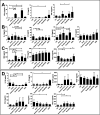
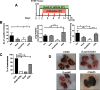
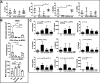
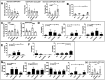
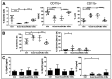
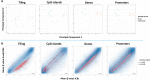
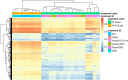
Results: In combination with ICBs, Guadecitabine significantly reduced subcutaneous tumor growth as well as metastases formation compared to ICBs and Guadecitabine treatment. In particular, Guadecitabine greatly enhanced the efficacy of combined ICBs by increasing effector memory CD8+ T cells, inducing effector NK cells in the spleen and reducing tumor infiltrating regulatory T cells and myeloid derived suppressor cells (MDSC), in the tumor microenvironment (TME). Guadecitabine in association with ICBs increased serum levels of IFN-γ and IFN-γ-induced chemokines with anti-angiogenic activity. Guadecitabine led to a general DNA-demethylation, in particular of sites of intermediate methylation levels.
Conclusions: These results indicate Guadecitabine as a promising epigenetic drug to be added to ICBs therapy.
Keywords: Anti-CTLA-4; Anti-PD-1; Guadecitabine; MDSC; Melanoma; Treg; Tumor microenvironment.
© 2023. The Author(s).
Conflict of interest statement
Michele Maio has served as a consultant and/or advisor to Roche, Bristol-Myers Squibb, Merck Sharp Dohme, Incyte, AstraZeneca, Amgen, Pierre Fabre, Eli Lilly, Glaxo Smith Kline, Sciclone, Sanofi, Alfasigma, and Merck Serono; and own shares in Theravance and Epigen Therapeutics, Srl. The other authors declare that they have no competing interests.
Figures



Similar articles
-
Future perspectives in melanoma research : Meeting report from the "Melanoma Bridge". Napoli, December 1st-4th 2015.J Transl Med. 2016 Nov 15;14(1):313. doi: 10.1186/s12967-016-1070-y. J Transl Med. 2016. PMID: 27846884 Free PMC article.
-
Targeting interferon signaling and CTLA-4 enhance the therapeutic efficacy of anti-PD-1 immunotherapy in preclinical model of HPV+ oral cancer.J Immunother Cancer. 2019 Sep 18;7(1):252. doi: 10.1186/s40425-019-0728-4. J Immunother Cancer. 2019. PMID: 31533840 Free PMC article.
-
The DNA methyltransferase inhibitor, guadecitabine, targets tumor-induced myelopoiesis and recovers T cell activity to slow tumor growth in combination with adoptive immunotherapy in a mouse model of breast cancer.BMC Immunol. 2020 Feb 27;21(1):8. doi: 10.1186/s12865-020-0337-5. BMC Immunol. 2020. PMID: 32106810 Free PMC article.
-
Immune checkpoint blockade in melanoma: Advantages, shortcomings and emerging roles of the nanoparticles.Int Immunopharmacol. 2022 Dec;113(Pt A):109300. doi: 10.1016/j.intimp.2022.109300. Epub 2022 Oct 17. Int Immunopharmacol. 2022. PMID: 36252486 Review.
-
Mechanisms of Resistance to Checkpoint Blockade Therapy.Adv Exp Med Biol. 2020;1248:83-117. doi: 10.1007/978-981-15-3266-5_5. Adv Exp Med Biol. 2020. PMID: 32185708 Review.
Cited by
-
The epigenetic hallmarks of immune cells in cancer.Mol Cancer. 2025 Mar 5;24(1):66. doi: 10.1186/s12943-025-02255-4. Mol Cancer. 2025. PMID: 40038722 Free PMC article. Review.
-
Recent advances on gene-related DNA methylation in cancer diagnosis, prognosis, and treatment: a clinical perspective.Clin Epigenetics. 2025 May 5;17(1):76. doi: 10.1186/s13148-025-01884-2. Clin Epigenetics. 2025. PMID: 40325471 Free PMC article. Review.
-
Targeting novel regulated cell death: disulfidptosis in cancer immunotherapy with immune checkpoint inhibitors.Biomark Res. 2025 Feb 26;13(1):35. doi: 10.1186/s40364-025-00748-4. Biomark Res. 2025. PMID: 40012016 Free PMC article. Review.
-
Dysregulated gene expression of SUMO machinery components induces the resistance to anti-PD-1 immunotherapy in lung cancer by upregulating the death of peripheral blood lymphocytes.Front Immunol. 2024 Aug 15;15:1424393. doi: 10.3389/fimmu.2024.1424393. eCollection 2024. Front Immunol. 2024. PMID: 39211047 Free PMC article.
-
Metastasis organotropism in colorectal cancer: advancing toward innovative therapies.J Transl Med. 2023 Sep 9;21(1):612. doi: 10.1186/s12967-023-04460-5. J Transl Med. 2023. PMID: 37689664 Free PMC article. Review.
References
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

