Inter-axonal molecular crosstalk via Lumican proteoglycan sculpts murine cervical corticospinal innervation by distinct subpopulations
- PMID: 36934325
- PMCID: PMC10167627
- DOI: 10.1016/j.celrep.2023.112182
Inter-axonal molecular crosstalk via Lumican proteoglycan sculpts murine cervical corticospinal innervation by distinct subpopulations
Abstract
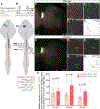
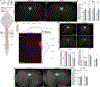
How CNS circuits sculpt their axonal arbors into spatially and functionally organized domains is not well understood. Segmental specificity of corticospinal connectivity is an exemplar for such regional specificity of many axon projections. Corticospinal neurons (CSN) innervate spinal and brainstem targets with segmental precision, controlling voluntary movement. Multiple molecularly distinct CSN subpopulations innervate the cervical cord for evolutionarily enhanced precision of forelimb movement. Evolutionarily newer CSNBC-lat exclusively innervate bulbar-cervical targets, while CSNmedial are heterogeneous; distinct subpopulations extend axons to either bulbar-cervical or thoraco-lumbar segments. We identify that Lumican controls balance of cervical innervation between CSNBC-lat and CSNmedial axons during development, which is maintained into maturity. Lumican, an extracellular proteoglycan expressed by CSNBC-lat, non-cell-autonomously suppresses cervical collateralization by multiple CSNmedial subpopulations. This inter-axonal molecular crosstalk between CSN subpopulations controls murine corticospinal circuitry refinement and forelimb dexterity. Such crosstalk is generalizable beyond the corticospinal system for evolutionary incorporation of new neuron populations into preexisting circuitry.
Keywords: CP: Neuroscience; Lumican proteoglycan; axon collateralization; axon development; circuit refinement; corticospinal neurons; corticospinal segmental specificity; evolutionary circuit diversification; forelimb dexterity; inter-axonal molecular crosstalk.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




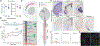
References
-
- Sahni V, Engmann A, Ozkan A, and Macklis JD (2020). Motor Cortex Connections, 2nd Edition (Elsevier; ).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

