Reference gene selection for clinical chimeric antigen receptor T-cell product vector copy number assays
- PMID: 36935289
- PMCID: PMC10159958
- DOI: 10.1016/j.jcyt.2023.02.010
Reference gene selection for clinical chimeric antigen receptor T-cell product vector copy number assays
Abstract
Background aims: Reference genes are an essential part of clinical assays such as droplet digital polymerase chain reaction (ddPCR), which measure the number of copies of vector integrated into genetically engineered cells and the loss of plasmids in reprogrammed cells used in clinical cell therapies. Care should be taken to select reference genes, because it has been discovered that there may be thousands of variations in copy number from genomic segments among different individuals. In addition, within the same person in the context of cancer and other proliferative disorders, substantial parts of the genome also can differ in copy number between cells from diseased and healthy people. The purpose of this study was to identify reference genes that could be used for copy number variation analysis of transduced chimeric antigen receptor T cells and for plasmid loss analysis in induced pluripotent stem cells using ddPCR.
Methods: We used The Cancer Genome Atlas (TCGA) to evaluate candidate reference genes. If TCGA found a candidate gene to have low copy number variance in cancer, ddPCR was used to measure the copy numbers of the potential reference gene in cells from healthy subjects, cancer cell lines and patients with acute lymphocytic leukemia, lymphoma, multiple myeloma and human papillomavirus-associated cancers.
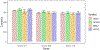
Results: In addition to the rPP30 gene, which we have has been using in our copy number assays, three other candidate reference genes were evaluated using TCGA, and this analysis found that none of the four gene regions (AGO1, AP3B1, MKL2 and rPP30) were amplified or deleted in all of the cancer cell types that are currently being treated with cellular therapies by our facility. The number of copies of the genes AP3B1, AGO1, rPP30 and MKL2 measured by ddPCR was similar among cells from healthy subjects. We found that AGO1 had copy number alteration in some of the clinical samples, and the number of copies of the genes AP3B1, MKL2 and rPP30 measured by ddPCR was similar among cells from patients with the cancer cell types that are currently being treated with genetically engineered T-cell therapies by our facility.
Conclusions: Based on our current results, the three genes, AP3B1, MKL2 and rPP30, are suitable for use as reference genes for assays measuring vector copy number in chimeric antigen receptor T cells produced from patients with acute leukemia, lymphoma, multiple myeloma and human papillomavirus-associated cancers. We will continue to evaluate AGO1 on our future samples.
Keywords: chimeric antigen receptor T-cells; droplet digital PCR; vector copy number.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest The authors have no commercial, proprietary or financial interest in the products or companies described in this article.
Figures


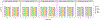
References
-
- Scheuermann RH, Racila E. CD19 antigen in leukemia and lymphoma diagnosis and immunotherapy. Leuk Lymphoma 1995;18:385–97. - PubMed
-
- Kochenderfer JN, Wilson WH, Janik JE, Dudley ME, Stetler-Stevenson M, Feldman SA, Maric I, Raffeld M, Nathan DA, Lanier BJ, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 2010;116:4099–102. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

