Ribonuclease inhibitor 1 (RNH1) deficiency cause congenital cataracts and global developmental delay with infection-induced psychomotor regression and anemia
- PMID: 36935417
- PMCID: PMC10400601
- DOI: 10.1038/s41431-023-01327-7
Ribonuclease inhibitor 1 (RNH1) deficiency cause congenital cataracts and global developmental delay with infection-induced psychomotor regression and anemia
Abstract
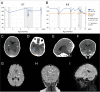
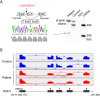
Ribonuclease inhibitor 1, also known as angiogenin inhibitor 1, encoded by RNH1, is a ubiquitously expressed leucine-rich repeat protein, which is highly conserved in mammalian species. Inactivation of rnh1 in mice causes an embryonically lethal anemia, but the exact biological function of RNH1 in humans remains unknown and no human genetic disease has so far been associated with RNH1. Here, we describe a family with two out of seven siblings affected by a disease characterized by congenital cataract, global developmental delay, myopathy and psychomotor deterioration, seizures and periodic anemia associated with upper respiratory tract infections. A homozygous splice-site variant (c.615-2A > C) in RNH1 segregated with the disease. Sequencing of RNA derived from patient fibroblasts and cDNA analysis of skeletal muscle mRNA showed aberrant splicing with skipping of exon 7. Western blot analysis revealed a total lack of the RNH1 protein. Functional analysis revealed that patient fibroblasts were more sensitive to RNase A exposure, and this phenotype was reversed by transduction with a lentivirus expressing RNH1 to complement patient cells. Our results demonstrate that loss-of-function of RNH1 in humans is associated with a multiorgan developmental disease with recessive inheritance. It may be speculated that the infection-induced deterioration resulted from an increased susceptibility toward extracellular RNases and/or other inflammatory responses normally kept in place by the RNase inhibitor RNH1.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Human molecular genetics sheds light on the physiological significance of ribonuclease inhibitor (RNH1).Eur J Hum Genet. 2023 Aug;31(8):856-858. doi: 10.1038/s41431-023-01362-4. Epub 2023 Apr 21. Eur J Hum Genet. 2023. PMID: 37085604 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

