CLDN6 inhibits breast cancer metastasis through WIP-dependent actin cytoskeleton-mediated autophagy
- PMID: 36935496
- PMCID: PMC10026481
- DOI: 10.1186/s13046-023-02644-x
CLDN6 inhibits breast cancer metastasis through WIP-dependent actin cytoskeleton-mediated autophagy
Abstract
Background: As a breast cancer suppressor gene, CLDN6 overexpression was found to inhibit breast cancer metastasis in our previous studies, but the specific mechanism remains unclear. This study aimed to clarify the role and mechanism of CLDN6 in inhibiting breast cancer metastasis.
Methods: Western blot, immunofluorescence and transmission electron microscopy were performed to detect autophagy. Wound healing, transwell assays and lung metastasis mouse models were used to examine breast cancer metastasis. Phalloidin staining and immunofluorescent staining were used to observe actin cytoskeleton. mRNA seq, RT-PCR, western blot, chromatin immunoprecipitation, dual luciferase reporter assay, co-immunoprecipitation and immunofluorescence were performed to define the molecular mechanism. The expression levels and clinical implication of CLDN6, WIP and LC3 in breast cancer tissues were evaluated using immunohistochemistry.
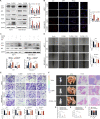
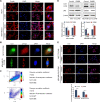
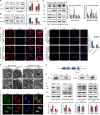
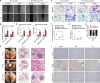
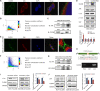
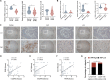
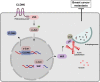
Results: We demonstrated that CLDN6 inhibited breast cancer metastasis through autophagy in vitro and vivo. We unraveled a novel mechanism that CLDN6 regulated autophagy via WIP-dependent actin cytoskeleton assembly. Through its PDZ-binding motif, overexpressed CLDN6 interacted with JNK and upregulated JNK/c-Jun pathway. C-Jun promoted WIP expression at the transcriptional level. Notably, we observed c-Jun transcriptionally upregulated CLDN6 expression, and there was a positive feedback loop between CLDN6 and JNK/c-Jun. Finally, we found that CLDN6, WIP and LC3 expression correlated with each other, and WIP expression was significantly associated with lymph node metastasis of breast cancer patients.
Conclusions: The data provide a new insight into the inhibitory effects of CLDN6-mediated autophagy on breast cancer metastasis, and revealed the new mechanism of CLDN6 regulating autophagy through WIP-dependent actin cytoskeleton. Our findings enrich the theoretical basis for CLDN6 as a potential biomarker for breast cancer diagnosis and therapy.
Keywords: Actin cytoskeleton; Autophagy; Breast cancer; CLDN6; Metastasis; WIP.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

