In vitro and in silico evaluations of actinomycin X2and actinomycin D as potent anti-tuberculosis agents
- PMID: 36935926
- PMCID: PMC10022501
- DOI: 10.7717/peerj.14502
In vitro and in silico evaluations of actinomycin X2and actinomycin D as potent anti-tuberculosis agents
Abstract
Background: Multidrug-resistant tuberculosis (MDR-TB) is one of the world's most devastating contagious diseases and is caused by the MDR-Mycobacterium tuberculosis (MDR-Mtb) bacteria. It is therefore essential to identify novel anti-TB drug candidates and target proteins to treat MDR-TB. Here, in vitro and in silico studies were used to investigate the anti-TB potential of two newly sourced actinomycins, actinomycin-X2 (act-X2) and actinomycin-D (act-D), from the Streptomyces smyrnaeus strain UKAQ_23 (isolated from the Jubail industrial city of Saudi Arabia).
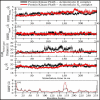
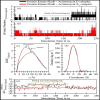
Methods: The anti-TB activity of the isolated actinomycins was assessed in vitro using the Mtb H37Ra, Mycobacterium bovis (BCG), and Mtb H37Rv bacterial strains, using the Microplate Alamar Blue Assay (MABA) method. In silico molecular docking studies were conducted using sixteen anti-TB drug target proteins using the AutoDock Vina 1.1.2 tool. The molecular dynamics (MD) simulations for both actinomycins were then performed with the most suitable target proteins, using the GROningen MAchine For Chemical Simulations (GROMACS) simulation software (GROMACS 2020.4), with the Chemistry at HARvard Macromolecular Mechanics 36m (CHARMM36m) forcefield for proteins and the CHARMM General Force Field (CGenFF) for ligands.
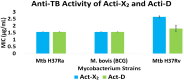
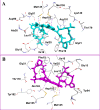
Results: In vitro results for the Mtb H37Ra, BCG, and Mtb H37Rv strains showed that act-X2 had minimum inhibitory concentration (MIC) values of 1.56 ± 0.0, 1.56 ± 0.0, and 2.64 ± 0.07 µg/mL and act-D had MIC values of 1.56 ± 0.0, 1.56 ± 0.0, and 1.80 ± 0.24 µg/mL respectively. The in silico molecular docking results showed that protein kinase PknB was the preferred target for both actinomycins, while KasA and pantothenate synthetase were the least preferred targets for act-X2and act-D respectively. The molecular dynamics (MD) results demonstrated that act-X2 and act-D remained stable inside the binding region of PknB throughout the simulation period. The MM/GBSA (Molecular Mechanics/Generalized Born Surface Area) binding energy calculations showed that act-X2 was more potent than act-D.
Conclusion: In conclusion, our results suggest that both actinomycins X2 and D are highly potent anti-TB drug candidates. We show that act-X2is better able to antagonistically interact with the protein kinase PknB target than act-D, and thus has more potential as a new anti-TB drug candidate.
Keywords: Actinomycin; Anti-TB drug; Molecular docking; Protein kinase PknB; Streptomyces.
©2023 Qureshi et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindah E. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. - DOI
-
- Almeleebia TM, AlShahrani M, Alshahrani MY, Ahmad I, Alkahtani AM, Alam MJ, Kausar MA, Saeed A, Saeed M, Iram S. Identification of new mycobacterium tuberculosis proteasome inhibitors using a knowledge-based computational screening approach. Molecules. 2021;26:2326. doi: 10.3390/molecules26082326. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

