Label-free cell segmentation of diverse lymphoid tissues in 2D and 3D
- PMID: 36936072
- PMCID: PMC10014308
- DOI: 10.1016/j.crmeth.2023.100398
Label-free cell segmentation of diverse lymphoid tissues in 2D and 3D
Abstract
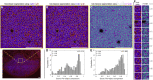
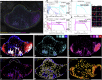
Unlocking and quantifying fundamental biological processes through tissue microscopy requires accurate, in situ segmentation of all cells imaged. Currently, achieving this is complex and requires exogenous fluorescent labels that occupy significant spectral bandwidth, increasing the duration and complexity of imaging experiments while limiting the number of channels remaining to address the study's objectives. We demonstrate that the excitation light reflected during routine confocal microscopy contains sufficient information to achieve accurate, label-free cell segmentation in 2D and 3D. This is achieved using a simple convolutional neural network trained to predict the probability that reflected light pixels belong to either nucleus, cytoskeleton, or background classifications. We demonstrate the approach across diverse lymphoid tissues and provide video tutorials demonstrating deployment in Python and MATLAB or via standalone software for Windows.
Keywords: 2D; 3D; cell segmentation; confocal microscopy; digital pathology; immunofluorescence; label free; quantitative; single-cell; tissue.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Stoltzfus C.R., Filipek J., Gern B.H., Olin B.E., Leal J.M., Wu Y., Lyons-Cohen M.R., Huang J.Y., Paz-Stoltzfus C.L., Plumlee C.R., et al. CytoMAP: a spatial analysis toolbox reveals features of myeloid cell organization in lymphoid tissues. Cell Rep. 2020;31:107523. doi: 10.1016/j.celrep.2020.107523. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

