Phylostems: a new graphical tool to investigate temporal signal of heterochronous sequences datasets
- PMID: 36936370
- PMCID: PMC10017117
- DOI: 10.1093/bioadv/vbad026
Phylostems: a new graphical tool to investigate temporal signal of heterochronous sequences datasets
Abstract
Motivation: Molecular tip-dating of phylogenetic trees is a growing discipline that uses DNA sequences sampled at different points in time to co-estimate the timing of evolutionary events with rates of molecular evolution. Importantly, such inferences should only be performed on datasets displaying sufficient temporal signal, a feature important to test prior to any tip-dating inference. For this purpose, the most popular method considered to-date has been the 'root-to-tip regression' which consist in fitting a linear regression of the number of substitutions accumulated from the root to the tips of a phylogenetic tree as a function of sampling times. The main limitation of the regression method, in its current implementation, relies in the fact that the temporal signal can only be tested at the whole-tree scale (i.e. its root).
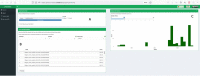
Results: To overcome this limitation we introduce Phylostems, a new graphical user-friendly tool developed to investigate temporal signal within every clade of a phylogenetic tree. We provide a 'how to' guide by running Phylostems on an empirical dataset and supply guidance for results interpretation.
Availability and implementation: Phylostems is freely available at https://pvbmt-apps.cirad.fr/apps/phylostems.
© The Author(s) 2023. Published by Oxford University Press.
Figures


References
-
- Chang W. et al. (2018) shiny: Web Application Framework for R. Comprehensive R Archive Network (CRAN). https://cran.r-project.org/package=shiny
-
- Drummond A.J. et al. (2003a) Inference of viral evolutionary rates from molecular sequences. Adv. Parasitol., 54, 331–358. - PubMed
-
- Drummond A.J. et al. (2003b). Measurably evolving populations. Trends Ecol. Evol., 18, 481–488.
LinkOut - more resources
Full Text Sources

