Complete chloroplast genome data of Shorea macrophylla (Engkabang): Structural features, comparative and phylogenetic analysis
- PMID: 36936629
- PMCID: PMC10018430
- DOI: 10.1016/j.dib.2023.109029
Complete chloroplast genome data of Shorea macrophylla (Engkabang): Structural features, comparative and phylogenetic analysis
Abstract
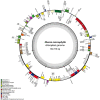
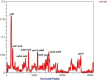
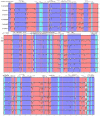
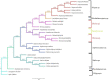
Shorea macrophylla belongs to the Shorea genus under the Dipterocarpaceae family. It is a woody tree that grows in the rainforest in Southeast Asia. The complete chloroplast (cp) genome sequence of S. macrophylla is reported here. The genomic size of S. macrophylla is 150,778 bp and it possesses a circular structure with conserved constitute regions of large single copy (LSC, 83,681 bp) and small single copy (SSC, 19,813 bp) regions, as well as a pair of inverted repeats with a length of 23,642 bp. It has 112 unique genes, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. The genome exhibits a similar GC content, gene order, structure, and codon usage when compared to previously reported chloroplast genomes from other plant species. The chloroplast genome of S. macrophylla contained 262 SSRs, the most prevalent of which was A/T, followed by AAT/ATT. Furthermore, the sequences contain 43 long repeat sequences, practically most of them are forward or palindrome type long repeats. The genome structure of S. macrophylla was compared to the genomic structures of closely related species from the same family, and eight mutational hotspots were discovered. The phylogenetic analysis demonstrated a close relationship between Shorea and Parashorea species, indicating that Shorea is not monophyletic. The complete chloroplast genome sequence analysis of S. macrophylla reported in this paper will contribute to further studies in molecular identification, genetic diversity, and phylogenetic research.
Keywords: Chloroplast genome; Dipterocarpaceae; Monophyletic; Phylogenetic analysis; Shorea macrophylla.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Chai E.O.K. University of Edinburgh; Malaysia: 1998. Aspects of a Tree Improvement Programme for Shorea Macrophylla (de Vriese) Ashton in Sarawak.
-
- Ashton P.S. Dipterocarpaceae. Flora Malesiana. 1982;9:237–552.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

