Strategies for optimizing CITE-seq for human islets and other tissues
- PMID: 36936943
- PMCID: PMC10014726
- DOI: 10.3389/fimmu.2023.1107582
Strategies for optimizing CITE-seq for human islets and other tissues
Abstract
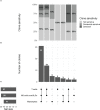
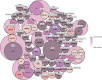
Defining the immunological landscape of human tissue is an important area of research, but challenges include the impact of tissue disaggregation on cell phenotypes and the low abundance of immune cells in many tissues. Here, we describe methods to troubleshoot and standardize Cellular Indexing of Transcriptomes and Epitopes by sequencing (CITE-seq) for studies involving enzymatic digestion of human tissue. We tested epitope susceptibility of 92 antibodies commonly used to differentiate immune lineages and cell states on human peripheral blood mononuclear cells following treatment with an enzymatic digestion cocktail used to isolate islets. We observed CD4, CD8a, CD25, CD27, CD120b, CCR4, CCR6, and PD1 display significant sensitivity to enzymatic treatment, effects that often could not be overcome with alternate antibodies. Comparison of flow cytometry-based CITE-seq antibody titrations and sequencing data supports that for the majority of antibodies, flow cytometry accurately predicts optimal antibody concentrations for CITE-seq. Comparison by CITE-seq of immune cells in enzymatically digested islet tissue and donor-matched spleen not treated with enzymes revealed little digestion-induced epitope cleavage, suggesting increased sensitivity of CITE-seq and/or that the islet structure may protect resident immune cells from enzymes. Within islets, CITE-seq identified immune cells difficult to identify by transcriptional signatures alone, such as distinct tissue-resident T cell subsets, mast cells, and innate lymphoid cells (ILCs). Collectively this study identifies strategies for the rational design and testing of CITE-seq antibodies for single-cell studies of immune cells within islets and other tissues.
Keywords: CITE-seq; flow cytometry; pancreas; single cell RNA seq; tissue immunity.
Copyright © 2023 Colpitts, Budd, Monajemi, Reid, Murphy, Ivison, Verchere, Levings and Crome.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

