sRNA expedites polycistronic mRNA decay in Escherichia coli
- PMID: 36936984
- PMCID: PMC10020718
- DOI: 10.3389/fmolb.2023.1097609
sRNA expedites polycistronic mRNA decay in Escherichia coli
Abstract
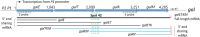
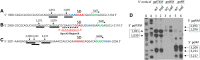
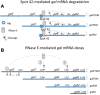
In bacteria, most small RNA (sRNA) elicits RNase E-mediated target mRNA degradation by binding near the translation initiation site at the 5' end of the target mRNA. Spot 42 is an sRNA that binds in the middle of the gal operon near the translation initiation site of galK, the third gene of four, but it is not clear whether this binding causes degradation of gal mRNA. In this study, we measured the decay rate of gal mRNA using Northern blot and found that Spot 42 binding caused degradation of only a specific group of gal mRNA that shares their 3' end with full-length mRNA. The results showed that in the MG1655Δspf strain in which the Spot 42 gene was removed, the half-life of each gal mRNA in the group increased by about 200% compared to the wild type. Since these mRNA species are intermediate mRNA molecules created by the decay process of the full-length gal mRNA, these results suggest that sRNA accelerates the mRNA decaying processes that normally operate, thus revealing an unprecedented role of sRNA in mRNA biology.
Keywords: RNase E; Spot42; galactose operon; mRNA decay; polarity; sRNA.
Copyright © 2023 Jeon, Lee, N, Kang and Lim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

