The rise of animal biotelemetry and genetics research data integration
- PMID: 36937069
- PMCID: PMC10019913
- DOI: 10.1002/ece3.9885
The rise of animal biotelemetry and genetics research data integration
Abstract
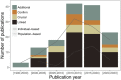
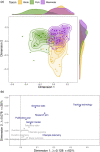
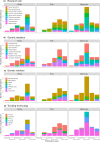
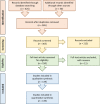
The advancement and availability of innovative animal biotelemetry and genomic technologies are improving our understanding of how the movements of individuals influence gene flow within and between populations and ultimately drive evolutionary and ecological processes. There is a growing body of work that is integrating what were once disparate fields of biology, and here, we reviewed the published literature up until January 2023 (139 papers) to better understand the drivers of this research and how it is improving our knowledge of animal biology. The review showed that the predominant drivers for this research were as follows: (1) understanding how individual-based movements affect animal populations, (2) analyzing the relationship between genetic relatedness and social structuring, and (3) studying how the landscape affects the flow of genes, and how this is impacted by environmental change. However, there was a divergence between taxa as to the most prevalent research aim and the methodologies applied. We also found that after 2010 there was an increase in studies that integrated the two data types using innovative statistical techniques instead of analyzing the data independently using traditional statistics from the respective fields. This new approach greatly improved our understanding of the link between the individual, the population, and the environment and is being used to better conserve and manage species. We discuss the challenges and limitations, as well as the potential for growth and diversification of this research approach. The paper provides a guide for researchers who wish to consider applying these disparate disciplines and advance the field.
Keywords: animal tracking; biotelemetry; genetics; genomics; individual‐based movement; movement ecology.
© 2023 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Figures


References
-
- Abdi, H. (2007). Discriminant correspondence analysis. In Salkind N. (Ed.), Encyclopedia of measurement and statistics. SAGE Publications.
-
- Bartolommei, P. , Gasperini, S. , Manzo, E. , Natali, C. , Ciofi, C. , & Cozzolino, R. (2016). Genetic relatedness affects socio‐spatial organization in a solitary carnivore, the European pine marten. Hystrix, 27, 1–3. 10.4404/hystrix-27.2-11876 - DOI
-
- Bazzi, G. , Ambrosini, R. , Caprioli, M. , Costanzo, A. , Liechti, F. , Gatti, E. , Gianfranceschi, L. , Podofillini, S. , Romano, A. , Romano, M. , Scandolara, C. , Saino, N. , & Rubolini, D. (2015). Clock gene polymorphism and scheduling of migration: A geolocator study of the barn swallow Hirundo rustica . Scientific Reports, 5, 1–7. 10.1038/srep12443 - DOI - PMC - PubMed
-
- Beaton, D. , Rieck, J. , Fatt, C. R. C. , & Abdi, H. (2019). Package “TExPosition” .
Publication types
Associated data
LinkOut - more resources
Full Text Sources

