First trans-eunicellane terpene synthase in bacteria
- PMID: 36937101
- PMCID: PMC10022577
- DOI: 10.1016/j.chempr.2022.12.006
First trans-eunicellane terpene synthase in bacteria
Abstract
Terpenoids are the largest family of natural products, but prokaryotes are vastly underrepresented in this chemical space. However, genomics supports vast untapped biosynthetic potential for terpenoids in bacteria. We discovered the first trans-eunicellane terpene synthase (TS), AlbS from Streptomyces albireticuli NRRL B-1670, in nature. Mutagenesis, deuterium labeling studies, and quantum chemical calculations provided extensive support for its cyclization mechanism. In addition, parallel stereospecific labeling studies with Bnd4, a cis-eunicellane TS, revealed a key mechanistic distinction between these two enzymes. AlbS highlights bacteria as a valuable source of novel terpenoids, expands our understanding of the eunicellane family of natural products and the enzymes that biosynthesize them, and provides a model system to address fundamental questions about the chemistry of 6,10-bicyclic ring systems.
Keywords: Bacterial terpenoids; diterpenoid; enzymes; eunicellane; genome mining; isotope labeling; mechanism; quantum chemical calculations; terpene synthase.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures
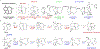

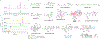


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
