Identification of iron metabolism-related genes in the circulation and myocardium of patients with sepsis via applied bioinformatics analysis
- PMID: 36937929
- PMCID: PMC10017502
- DOI: 10.3389/fcvm.2023.1018422
Identification of iron metabolism-related genes in the circulation and myocardium of patients with sepsis via applied bioinformatics analysis
Abstract
Background: Early diagnosis of septic cardiomyopathy is essential to reduce the mortality rate of sepsis. Previous studies indicated that iron metabolism plays a vital role in sepsis-induced cardiomyopathy. Here, we aimed to identify shared iron metabolism-related genes (IMRGs) in the myocardium and blood monocytes of patients with sepsis and to determine their prognostic signature.
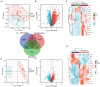
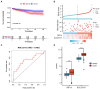
Methods: First, an applied bioinformatics-based analysis was conducted to identify shared IMRGs differentially expressed in the myocardium and peripheral blood monocytes of patients with sepsis. Second, Cytoscape was used to construct a protein-protein interaction network, and immune infiltration of the septic myocardium was assessed using single-sample gene set enrichment analysis. In addition, a prognostic prediction model for IMRGs was established by Cox regression analysis. Finally, the expression of key mRNAs in the myocardium of mice with sepsis was verified using quantitative polymerase chain reaction analysis.
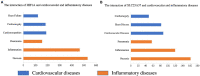
Results: We screened common differentially expressed genes in septic myocardium and blood monocytes and identified 14 that were related to iron metabolism. We found that HBB, SLC25A37, SLC11A1, and HMOX1 strongly correlated with monocytes and neutrophils, whereas HMOX1 and SLC11A1 strongly correlated with macrophages. We then established a prognostic model (HIF1A and SLC25A37) using the common differentially expressed IMRGs. The prognostic model we established was expected to better aid in diagnosing septic cardiomyopathy. Moreover, we verified these genes using datasets and experiments and found a significant difference between the sepsis and control groups.
Conclusion: Common differential expression of IMRGs was identified in blood monocytes and myocardium between sepsis and control groups, among which HIF1A and SLC25A37 might predict prognosis in septic cardiomyopathy. The study may help us deeply understand the molecular mechanisms of iron metabolism and aid in the diagnosis and treatment of septic cardiomyopathy.
Keywords: diagnostic biomarkers; iron metabolism; peripheral blood monocytes; sepsis; septic cardiomyopathy.
Copyright © 2023 Zhang, Di, Gao, Zhu, Li, Zhu, Wang, Wang, Zhou and Wang.
Conflict of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

