Genome-wide characterization and identification of Trihelix transcription factors and expression profiling in response to abiotic stresses in Chinese Willow (Salix matsudana Koidz)
- PMID: 36938039
- PMCID: PMC10020544
- DOI: 10.3389/fpls.2023.1125519
Genome-wide characterization and identification of Trihelix transcription factors and expression profiling in response to abiotic stresses in Chinese Willow (Salix matsudana Koidz)
Abstract
Trihelix transcription factors (TTF) are a class of light-responsive proteins with a typical triple-helix structure (helix-loop-helix-loop-helix). Members of this gene family play an important role in plant growth and development, especially in various abiotic stress responses. Salix matsudana Koidz is an allotetraploid ornamental forest tree that is widely planted for its excellent resistance to stress, but no studies on its Trihelix gene family have been reported. In this study, the Trihelix gene family was analyzed at the genome-wide level in S. matsudana. A total of 78 S. matsudana Trihelix transcription factors (SmTTFs) were identified, distributed on 29 chromosomes, and classified into four subfamilies (GT-1, GT-2, SH4, SIP1) based on their structural features. The gene structures and conserved functional domains of these Trihelix genes are similar in the same subfamily and differ between subfamilies. The presence of multiple stress-responsive cis-elements on the promoter of the S. matsudana Trihelix gene suggests that the S. matsudana Trihelix gene may respond to abiotic stresses. Expression pattern analysis revealed that Trihelix genes have different functions during flooding stress, salt stress, drought stress and low temperature stress in S. matsudana. Given that SmTTF30, as a differentially expressed gene, has a faster response to flooding stress, we selected SmTTF30 for functional studies. Overexpression of SmTTF30 in Arabidopsis thaliana (Arabidopsis) enhances its tolerance to flooding stress. Under flooding stress, the leaf cell activity and peroxidase activity (POD) of the overexpression strain were significantly higher than the leaf cell activity and POD of the wild type, and the malondialdehyde (MDA) content was significantly lower than the MDA content of the wild type. Thus, these results suggest that SmTTF30 enhances plant flooding tolerance and plays a positive regulatory role in plant flooding tolerance.
Keywords: RNA-Seq; Salix matsudana; Trihelix family; genome-wide characterization; submergence stress.
Copyright © 2023 Yang, Tang, Yang, Huang, Wang, Huang, Wei, Liu, Lian, Chen and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Figures



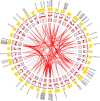
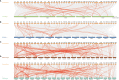




References
-
- Bailey T. L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

