Differentially methylated regions identified in bovine embryos are not observed in adulthood
- PMID: 36938311
- PMCID: PMC10023072
- DOI: 10.1590/1984-3143-AR2022-0076
Differentially methylated regions identified in bovine embryos are not observed in adulthood
Abstract
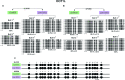
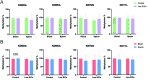
The establishment of epigenetic marks during the reprogramming window is susceptible to environmental influences, and stimuli during this critical stage can cause altered DNA methylation in offspring. In a previous study, we found that low levels of sulphur and cobalt (low S/Co) in the diet offered to oocyte donors altered the DNA methylome of bovine embryos. However, due to the extensive epigenetic reprogramming that occurs during embryogenesis, we hypothesized that the different methylation regions (DMRs) identified in the blastocysts may not maintain in adulthood. Here, we aimed to characterize DMRs previously identified in embryos, in the blood and sperm of adult progenies of two groups of heifers (low S/Co and control). We used six bulls and characterized the DNA methylation levels of KDM2A, KDM5A, KMT2D, and DOT1L genes. Our results showed that all DMRs analysed in both groups and tissues were hypermethylated unlike that noticed in the embryonic methylome profiles. These results suggest that embryo DMRs were reprogrammed during the final stages of de novo methylation during embryogenesis or later in development. Therefore, due to the highly dynamic epigenetic state during early embryonic development, we suggest that is essential to validate the DMRs found in embryos in adult individuals.
Keywords: DMRs; cattle; epigenetics; methylome; reprogramming.
Conflict of interest statement
Conflicts of interest: The authors have no conflict of interest to declare.
Figures




References
-
- Capra E, Lazzari B, Turri F, Cremonesi P, Portela AMR, Ajmone-Marsan P, Stella A, Pizzi F. Epigenetic analysis of high and low motile sperm populations reveals methylation variation in satellite regions within the pericentromeric position and in genes functionally related to sperm DNA organization and maintenance in Bos taurus. BMC Genomics. 2019;20(1):940. doi: 10.1186/s12864-019-6317-6. - DOI - PMC - PubMed
-
- Carlin G, Chaumontet C, Blachier F, Barbillon P, Darcel N, Blais A, Delteil C, Guillin FM, Blat S, van der Beek EM, Kodde A, Tome D, Davila AM. Maternal high-protein diet during pregnancy modifies rat offspring body weight and insulin signalling but not macronutrient preference in adulthood. Nutrients. 2019;11(1):96. doi: 10.3390/nu11010096. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
