Vancomycin Resistance in Enterococcus faecium from the Dallas, Texas, Area Is Conferred Predominantly on pRUM-Like Plasmids
- PMID: 36939336
- PMCID: PMC10117061
- DOI: 10.1128/msphere.00024-23
Vancomycin Resistance in Enterococcus faecium from the Dallas, Texas, Area Is Conferred Predominantly on pRUM-Like Plasmids
Abstract
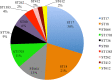
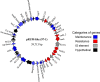
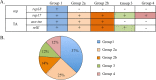
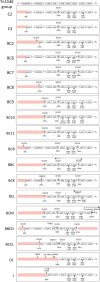
Vancomycin-resistant E. faecium (VREfm) is a significant public health concern because of limited treatment options. Genomic surveillance can be used to monitor VREfm transmission and evolution. Genomic analysis of VREfm has not been reported for the Dallas/Fort Worth/Arlington, TX, area, which is currently the 4th largest metropolitan area in the United States. Our study aimed to address this gap in knowledge by analyzing the genomes of 46 VREfm strains and 1 vancomycin-sensitive comparator collected during routine fecal surveillance of high-risk patients upon admission to a Dallas, TX, hospital system (August to October 2015). Thirty-one complete and 16 draft genome sequences were generated. The closed VREfm genomes possessed up to 12 extrachromosomal elements each. Overall, 251 closed putative plasmid sequences assigned to previously described and newly defined rep family types were obtained. Phylogenetic analysis identified 10 different sequence types (STs) among the isolates, with the most prevalent being ST17 and ST18. Strikingly, all but three of the VREfm isolates encoded vanA-type vancomycin resistance within Tn1546-like elements on a pRUM-like (rep17) plasmid backbone. Relative to a previously reported typing scheme for the vanA-carrying Tn1546, new variants of the Tn1546 were identified that harbored a combination of 7 insertion sequences (IS), including 3 novel IS elements reported here (ISEfa16, ISEfa17, and ISEfa18). We conclude that pRUM-like plasmids are important vectors for vancomycin resistance in the Dallas, TX, area and should be a focus of plasmid surveillance efforts. IMPORTANCE Vancomycin is an antibiotic used to treat infections caused by multidrug-resistant Gram-positive bacteria. Vancomycin resistance is common in clinical isolates of the Gram-positive pathogen Enterococcus faecium. Among E. faecium strains, vancomycin resistance genes can be disseminated by plasmids with different host ranges and transfer efficiencies. Surveillance of resistance plasmids is critical to understanding antibiotic resistance transmission. This study analyzed the genome sequences of VREfm isolates collected from the Dallas, TX, area, with particular focus on the mobile elements associated with vancomycin resistance genes. We found that a single plasmid family, the pRUM-like family, was associated with vancomycin resistance in the majority of isolates sampled. Our work suggests that the pRUM-like plasmids should continue to be studied to understand their mechanisms of maintenance, transmission, and evolution in VREfm.
Keywords: Enterococcus; mobile genetic element; mobile genetic elements; plasmid; plasmids; vancomycin resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00318-11. doi:10.1128/mBio.00318-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
