Harnessing the Noncanonical Keap1-Nrf2 Pathway for Human Cytomegalovirus Control
- PMID: 36939350
- PMCID: PMC10134830
- DOI: 10.1128/jvi.00160-23
Harnessing the Noncanonical Keap1-Nrf2 Pathway for Human Cytomegalovirus Control
Abstract
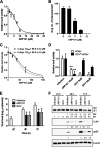
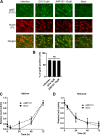
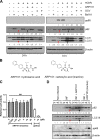
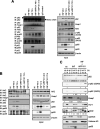
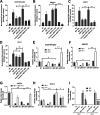
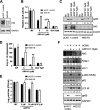
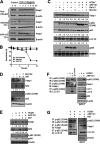
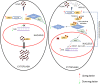
Host-derived cellular pathways can provide an unfavorable environment for virus replication. These pathways have been a subject of interest for herpesviruses, including the betaherpesvirus human cytomegalovirus (HCMV). Here, we demonstrate that a compound, ARP101, induces the noncanonical sequestosome 1 (SQSTM1)/p62-Keap1-Nrf2 pathway for HCMV suppression. ARP101 increased the levels of both LC3 II and SQSTM1/p62 and induced phosphorylation of p62 at the C-terminal domain, resulting in its increased affinity for Keap1. ARP101 treatment resulted in Nrf2 stabilization and translocation into the nucleus, binding to specific promoter sites and transcription of antioxidant enzymes under the antioxidant response element (ARE), and HCMV suppression. Knockdown of Nrf2 recovered HCMV replication following ARP101 treatment, indicating the role of the Keap1-Nrf2 axis in HCMV inhibition by ARP101. SQSTM1/p62 phosphorylation was not modulated by the mTOR kinase or casein kinase 1 or 2, indicating ARP101 engages other kinases. Together, the data uncover a novel antiviral strategy for SQSTM1/p62 through the noncanonical Keap1-Nrf2 axis. This pathway could be further exploited, including the identification of the responsible kinases, to define the biological events during HCMV replication. IMPORTANCE Antiviral treatment for human cytomegalovirus (HCMV) is limited and suffers from the selection of drug-resistant viruses. Several cellular pathways have been shown to modulate HCMV replication. The autophagy receptor sequestosome 1 (SQSTM1)/p62 has been reported to interact with several HCMV proteins, particularly with components of HCMV capsid, suggesting it plays a role in viral replication. Here, we report on a new and unexpected role for SQSTM1/p62, in HCMV suppression. Using a small-molecule probe, ARP101, we show SQSTM1/p62 phosphorylation at its C terminus domain initiates the noncanonical Keap1-Nrf2 axis, leading to transcription of genes under the antioxidant response element, resulting in HCMV inhibition in vitro. Our study highlights the dynamic nature of SQSTM1/p62 during HCMV infection and how its phosphorylation activates a new pathway that can be exploited for antiviral intervention.
Keywords: ARP101; SQSTM1/p62; antioxidant response element; human cytomegalovirus; noncanonical Keap1-Nrf2 pathway; p62-Keap1-Nrf2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kovacs A, Schluchter M, Easley K, Demmler G, Shearer W, La RP, Pitt J, Cooper E, Goldfarb J, Hodes D, Kattan M, McIntosh K. 1999. Cytomegalovirus infection and HIV-1 disease progression in infants born to HIV-1-infected women. Pediatric Pulmonary and Cardiovascular Complications of Vertically Transmitted HIV Infection Study Group. N Engl J Med 341:77–84. 10.1056/NEJM199907083410203. - DOI - PMC - PubMed
-
- Sabin CA, Phillips AN, Lee CA, Janossy G, Emery V, Griffiths PD. 1995. The effect of CMV infection on progression of human immunodeficiency virus disease is a cohort of haemophilic men followed for up to 13 years from seroconversion. Epidemiol Infect 114:361–372. 10.1017/s095026880005799x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

